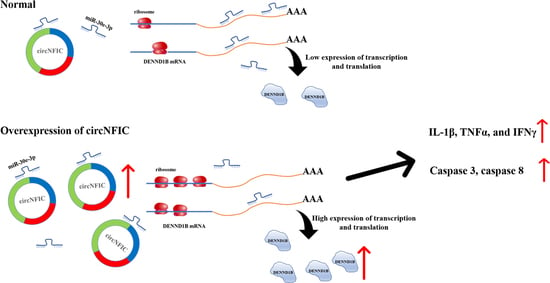
CircNFIC Balances Inflammation and Apoptosis by Sponging miR-30e-3p and Regulating DENND1B Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Construction of Cell Inflammatory Model
2.3. Circular Structure Confirmation
2.4. RNA Oligonucleotides used in This Study
2.5. Construction of Vectors
2.6. miRNA Targets Prediction and RNAhybrid Detection
2.7. Biotin Labeled Probe Pull-Down Assay
2.8. RNA Fluorescent In Situ Hybridization
2.9. RNA Isolation, Reverse Transcription
2.10. Quantitative Real-Time PCR and Western Blotting
2.11. Dual-Luciferase Reporter Assay
2.12. Statistical Analysis
3. Results
3.1. Characteristics and Subcellular Localization of circNFIC
3.2. circNFIC Aggravates Inflammation, Apoptosis, and Direct Targets miR-30e-3p
3.3. The gga-miR-30e-3p Suppresses Inflammation and Apoptosis and Direct Targets DENND1B
3.4. circNFIC Served as a Sponge of miR-30e-3p to Regulate the Expression of DENND1B
3.5. DENND1B-Induced Inflammation and Apoptosis in HD11
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scanes, C. Contribution of Poultry to Quality of Life and Economic Development in the Developing World. Poult. Sci. 2007, 86, 2289–2290. [Google Scholar] [CrossRef] [Green Version]
- Lawal, J.R.; Jajere, S.M.; Ibrahim, U.I.; Geidam, Y.A.; Gulani, I.A.; Musa, G.; Ibekwe, B.U. Prevalence of coccidiosis among village and exotic breed of chickens in Maiduguri, Nigeria. Veter World 2016, 9, 653–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Veter Res. 2020, 51, 115. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.B. A compartmentalised model for the estimation of the cost of coccidiosis to the world’s chicken production industry. Int. J. Parasitol. 1999, 29, 1209–1229. [Google Scholar] [CrossRef]
- McDonald, V.; Shirley, M.W. Past and future: Vaccination against Eimeria. Parasitology 2009, 136, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Kim, W.H.; Lillehoj, E.P.; Lillehoj, H.S. Recent progress in host immunity to avian coccidiosis: IL-17 family cytokines as sentinels of the intestinal mucosa. Dev. Comp. Immunol. 2013, 41, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Miska, K.B.; Kim, S.; Fetterer, R.H.; Dalloul, R.A.; Jenkins, M.C. Macrophage migration inhibitory factor (MIF) of the protozoan parasite Eimeria influences the components of the immune system of its host, the chicken. Parasitol. Res. 2013, 112, 1935–1944. [Google Scholar] [CrossRef]
- Kim, S.; Cox, C.M.; Jenkins, M.C.; Fetterer, R.H.; Miska, K.B.; Dalloul, R.A. Both host and parasite MIF molecules bind to chicken macrophages via CD74 surface receptor. Dev. Comp. Immunol. 2014, 47, 319–326. [Google Scholar] [CrossRef]
- Peek, H.; Landman, W.J. Coccidiosis in poultry: Anticoccidial products, vaccines and other prevention strategies. Veter Q. 2011, 31, 143–161. [Google Scholar] [CrossRef]
- Lee, S.-H.; Lillehoj, H.S.; Jang, S.I.; Hong, Y.-H.; Min, W.; Lillehoj, E.P.; Yancey, R.J.; Dominowski, P. Embryo vaccination of chickens using a novel adjuvant formulation stimulates protective immunity against Eimeria maxima infection. Vaccine 2010, 28, 7774–7778. [Google Scholar] [CrossRef]
- Dalloul, R.A.; Lillehoj, H.S. Recent Advances in Immunomodulation and Vaccination Strategies Against Coccidiosis. Avian Dis. 2005, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.B. Anticoccidial vaccines for broiler chickens: Pathways to success. Avian Pathol. 2002, 31, 317–353. [Google Scholar] [CrossRef]
- Pogonka, T.; Klotz, C.; Kovács, F.; Lucius, R. A single dose of recombinant Salmonella typhimurium induces specific humoral immune responses against heterologous Eimeria tenella antigens in chicken. Int. J. Parasitol. 2003, 33, 81–88. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Han, B.; Liu, H.; Wang, J.; Feng, X.; Sun, W.; Cai, D.; Jia, H.; Jiang, D. Circular RNA circACSL1 aggravated myocardial inflammation and myocardial injury by sponging miR-8055 and regulating MAPK14 expression. Cell Death Dis. 2021, 12, 487. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhang, J.; Ran, Y.; Pan, H.; Jia, J.; Zhao, Y.; Zhao, X.; Li, W.; Song, S.; Yu, X. Circular RNA hsa_circ_101555 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-145-5p and regulating CDCA3 expression. Cell Death Dis. 2021, 12, 356. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cheng, M.; Gu, B.; Wang, J.; Yan, S.; Xu, D. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 2020, 11, 833. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Li, X.; Zhang, M.; Lv, K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int. J. Mol. Med. 2017, 39, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon-Ming, C.; Marinov, G.K.; Chin, Y.-M.; Lim, Y.-Y.; Ea, C.-K. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci. Rep. 2017, 7, 12227. [Google Scholar] [CrossRef]
- Ng, W.L.; Marinov, G.K.; Liau, E.S.; Lam, Y.L.; Lim, Y.-Y.; Ea, C.-K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016, 13, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, S.; Zhang, Q.; Luo, Y.; Zhang, C.; Huan, Q.; Zhang, C. Overexpression of mm9_circ_013935 alleviates renal inflammation and fibrosis in diabetic nephropathy via the miR-153-3p/NFIC axis. Can. J. Physiol. Pharmacol. 2021, 99, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Dong, R.; Zhao, H. Inhibition of microRNA-346 inhibits myocardial inflammation and apoptosis after myocar-dial infarction via targeting NFIB. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11752–11760. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Su, B.; Wang, X.; Sun, Y.; Long, M.; Zheng, J.; Wu, W.; Li, L. miR-30e-3p Promotes Cardiomyocyte Autophagy and Inhibits Apoptosis via Regulating Egr-1 during Ischemia/Hypoxia. BioMed Res. Int. 2020, 2020, 7231243. [Google Scholar] [CrossRef] [PubMed]
- Pearson-Leary, J.; Eacret, D.; Chen, R.J.; Takano, H.; Nicholas, B.; Bhatnagar, S. Inflammation and vascular remodeling in the ventral hippocampus contributes to vulnerability to stress. Transl. Psychiatry 2017, 7, e1160. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Bradley, M.J.; Cai, Y.; Kummel, D.; De La Cruz, E.M.; Barr, F.; Reinisch, K.M. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl. Acad. Sci. USA 2011, 108, 18672–18677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marat, A.; Dokainish, H.; McPherson, P.S. DENN Domain Proteins: Regulators of Rab GTPases. J. Biol. Chem. 2011, 286, 13791–13800. [Google Scholar] [CrossRef] [Green Version]
- Shadrin, A.A.; Mucha, S.; Ellinghaus, D.; Makarious, M.B.; Blauwendraat, C.; Sreelatha, A.A.K.; Heras-Garvin, A.; Ding, J.; Hammer, M.; Foubert-Samier, A.; et al. Shared Genetics of Multiple System Atrophy and Inflammatory Bowel Disease. Mov. Disord. 2021, 36, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Godar, M.; Lambrecht, B.N. A New aDENNDum to Genetics of Childhood Asthma. Cell 2016, 164, 11–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, D.L.; Siminovitch, K.A.; Zamel, R.; Chapman, K.R.; Martin, N.G.; Zamel, N. Variation at DENND1B and Asthma on the Island of Tristan da Cunha. Twin Res. Hum. Genet. 2019, 22, 277–282. [Google Scholar] [CrossRef]
- Yang, C.-W.; Hojer, C.D.; Zhou, M.; Wu, X.; Wuster, A.; Lee, W.P.; Yaspan, B.; Chan, A.C. Regulation of T Cell Receptor Signaling by DENND1B in T H 2 Cells and Allergic Disease. Cell 2016, 164, 141–155. [Google Scholar] [CrossRef]
- Miyoshi, J.; Takai, Y. Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol. Med. 2004, 10, 476–480. [Google Scholar] [CrossRef]
- Bass, J.; Wilkinson, D.J.; Rankin, D.; Phillips, B.; Szewczyk, N.; Smith, K.; Atherton, P.J. An overview of technical considerations for Western blotting applications to physiological research. Scand. J. Med. Sci. Sports 2017, 27, 4–25. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Lv, H.; Yu, Q.; Zhou, Z.; Zhu, Q.; Wang, Z.; Cooper, P.; Smith, A.J.; Niu, Z.; et al. Human Stem Cells from the Apical Papilla Response to Bacterial Lipopolysaccharide Exposure and Anti-inflammatory Effects of Nuclear Factor I C. J. Endod. 2013, 39, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Calandra, T.; Bernhagen, J.; Mitchell, R.A.; Bucala, R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 1994, 179, 1895–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacher, M.; Metz, C.N.; Calandra, T.; Mayer, K.; Chesney, J.; Lohoff, M.; Gemsa, D.; Donnelly, T.; Bucala, R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc. Natl. Acad. Sci. USA 1996, 93, 7849–7854. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.B. Intercurrent coccidiosis and necrotic enteritis of chickens: Rational, integrated disease management by maintenance of gut integrity. Avian Pathol. 2005, 34, 159–180. [Google Scholar] [CrossRef]
- Blake, D.P.; Tomley, F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014, 30, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.H.; Chaudhari, A.A.; Lillehoj, H.S. Involvement of T Cell Immunity in Avian Coccidiosis. Front. Immunol. 2019, 10, 2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef]
- Miller, S.I.; Ernst, R.; Bader, M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005, 3, 36–46. [Google Scholar] [CrossRef]
- Jing, R.; Zhou, Z.; Kuang, F.; Huang, L.; Li, C. microRNA-99a Reduces Lipopolysaccharide-Induced Oxidative Injury by Activating Notch Pathway in H9c2 Cells. Int. Hearth J. 2017, 58, 422–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.L.G.; Salvesen, G.S. A primer on caspase mechanisms. Semin. Cell Dev. Biol. 2018, 82, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Gerner, C.; Fröhwein, U.; Gotzmann, J.; Bayer, E.; Gelbmann, D.; Bursch, W.; Schulte-Hermann, R. The Fas-induced Apoptosis Analyzed by High Throughput Proteome Analysis. J. Biol. Chem. 2000, 275, 39018–39026. [Google Scholar] [CrossRef] [Green Version]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 2018, 9, e1463. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Sun, H.; Shi, C.; Yang, H.; Wu, Y.; Li, W.; Dong, Y.; Cai, L.; Meng, X. Circular RNA in renal diseases. J. Cell. Mol. Med. 2020, 24, 6523–6533. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Lee, E.E.; Kim, J.; Yang, R.; Chamseddin, B.; Ni, C.; Gusho, E.; Xie, Y.; Chiang, C.-M.; Buszczak, M.; et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019, 10, 2300. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.-K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Hearth J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Li, Z.-Y.; Zhang, L.-M.; Wu, X.-Y.; Xiang, S.-H.; Wang, Y.-G.; Zhang, Y.-Q. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.S.; Lim, J.W.; Richards, L.J.; Bunt, J. The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. 2017, 410, 124–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, S.; Piper, M.; Gronostajski, R.M.; Richards, L.J. Nuclear Factor One Transcription Factors in CNS Development. Mol. Neurobiol. 2009, 39, 10–23. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, S.; Qi, Q.; Yang, X.; Zhu, E.; Yuan, H.; Li, X.; Liu, Y.; Li, X.; Wang, B. Nuclear factor I-C reciprocally regulates adipocyte and osteoblast differentiation via control of canonical Wnt signaling. FASEB J. 2017, 31, 1939–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idriss, H.T.; Naismith, J.H. TNF alpha and the TNF receptor superfamily: Structure-function relationship(s). Microsc. Res. Tech. 2000, 50, 184–195. [Google Scholar] [CrossRef]
- Delair, E.; Creuzet, C.; Dupouy-Camet, J.; Roisin, M.P. In vitro effect of TNF-alpha and IFN-gamma in retinal cell infec-tion with Toxoplasma gondii. Invest. Ophthalmol. Vis. Sci. 2009, 50, 1754–1760. [Google Scholar] [CrossRef]
- Gerondakis, S.; Grumont, R.J.; Gugasyan, R.; Wong, W.W.-L.; Isomura, I.; Ho, W.; Banerjee, A.K. Unravelling the complexities of the NF-KB signalling pathway using mouse knockout and transgenic models. Oncogene 2006, 25, 6781–6799. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFKB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstrepen, L.; Bekaert, T.; Chau, T.-L.; Tavernier, J.; Chariot, A.; Beyaert, R. TLR-4, IL-1R and TNF-R signaling to NF-KB: Variations on a common theme. Cell. Mol. Life Sci. 2008, 65, 2964–2978. [Google Scholar] [CrossRef]
- Baldwin, A.S. THE NF-KB AND IKB PROTEINS: New Discoveries and Insights. Annu. Rev. Immunol. 1996, 14, 649–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, Z.; Chen, X.; Peng, X.; Nie, Q. CircNFIC Balances Inflammation and Apoptosis by Sponging miR-30e-3p and Regulating DENND1B Expression. Genes 2021, 12, 1829. https://doi.org/10.3390/genes12111829
Chen Y, Wang Z, Chen X, Peng X, Nie Q. CircNFIC Balances Inflammation and Apoptosis by Sponging miR-30e-3p and Regulating DENND1B Expression. Genes. 2021; 12(11):1829. https://doi.org/10.3390/genes12111829
Chicago/Turabian StyleChen, Yangfeng, Zhijun Wang, Xiaolan Chen, Xi Peng, and Qinghua Nie. 2021. "CircNFIC Balances Inflammation and Apoptosis by Sponging miR-30e-3p and Regulating DENND1B Expression" Genes 12, no. 11: 1829. https://doi.org/10.3390/genes12111829
APA StyleChen, Y., Wang, Z., Chen, X., Peng, X., & Nie, Q. (2021). CircNFIC Balances Inflammation and Apoptosis by Sponging miR-30e-3p and Regulating DENND1B Expression. Genes, 12(11), 1829. https://doi.org/10.3390/genes12111829