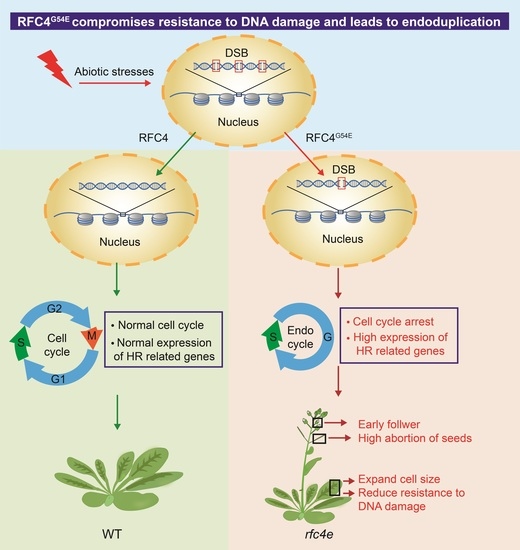
A Single Amino Acid Substitution in RFC4 Leads to Endoduplication and Compromised Resistance to DNA Damage in Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Construction of Binary Vectors and Transformation
2.3. RNA Extract and RT-qPCR
2.4. Transcriptome Analysis
2.5. True Leaf and Root Growth Inhibition Assays
2.6. Genotoxic Treatments
2.7. γ-H2AX Assays
2.8. Flow Cytometry
2.9. Leaf Epidermal Cell Examination
3. Results
3.1. rfc4e Mutants Were Generated by Site-Directed Mutagenesis
3.2. Mutation of AtRFC4 Causes Earlier Flowering and Seed Abortion
3.3. DNA Repair- and Replication-Related Genes Are Highly Upregulated in rfc4e Mutants
3.4. AtRFC4 Mutation Increases the Frequency of DNA Lesions
3.5. rfc4e Mutants Are Supersensitive to DSB-Inducing Genotoxic Agents
3.6. AtRFC4 Mutation Delays Cell Cycle and Promotes Endoduplication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsurimoto, T.; Stillman, B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell Biol. 1989, 9, 609–619. [Google Scholar] [CrossRef]
- Yao, N.Y.; O’Donnell, M. The RFC clamp loader: Structure and function. Subcell. Biochem. 2012, 62, 259–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majka, J.; Burgers, P.M. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid. Res. Mol. Biol. 2004, 78, 227–260. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, Y.; Nishitani, H. Control of Genome Integrity by RFC Complexes; Conductors of PCNA Loading onto and Unloading from Chromatin during DNA Replication. Genes (Basel) 2017, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Mossi, R.; Hubscher, U. Clamping down on clamps and clamp loaders--the eukaryotic replication factor C. Eur. J. Biochem. 1998, 254, 209–216. [Google Scholar] [CrossRef]
- Cullmann, G.; Fien, K.; Kobayashi, R.; Stillman, B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell Biol. 1995, 15, 4661–4671. [Google Scholar] [CrossRef] [Green Version]
- Podust, V.N.; Tiwari, N.; Ott, R.; Fanning, E. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J. Biol. Chem. 1998, 273, 12935–12942. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.L.; Pautz, A.L.; Burgers, P.M. ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J. Biol. Chem. 2001, 276, 34792–34800. [Google Scholar] [CrossRef] [Green Version]
- Uhlmann, F.; Cai, J.; Gibbs, E.; O’Donnell, M.; Hurwitz, J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J. Biol. Chem. 1997, 272, 10058–10064. [Google Scholar] [CrossRef] [Green Version]
- Uhlmann, F.; Gibbs, E.; Cai, J.; O’Donnell, M.; Hurwitz, J. Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J. Biol. Chem. 1997, 272, 10065–10071. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Qian, J.; You, L.; Zhang, X.; Jiao, J.; Liu, Y.; Zhao, J. Subunit Interaction Differences Between the Replication Factor C Complexes in Arabidopsis and Rice. Front. Plant. Sci. 2018, 9, 779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.; Chen, Y.; Hu, Y.; Deng, Y.; Liu, Y.; Li, G.; Zou, W.; Zhao, J. Arabidopsis replication factor C4 is critical for DNA replication during the mitotic cell cycle. Plant J. 2018, 94, 288–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckwith, W.H.; Sun, Q.; Bosso, R.; Gerik, K.J.; Burgers, P.M.; McAlear, M.A. Destabilized PCNA trimers suppress defective Rfc1 proteins in vivo and in vitro. Biochemistry 1998, 37, 3711–3722. [Google Scholar] [CrossRef] [PubMed]
- Noskov, V.N.; Araki, H.; Sugino, A. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol. Cell Biol. 1998, 18, 4914–4923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, K.; Ando, S.; Shimomura, T.; Matsumoto, K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell Biol. 1997, 17, 5905–5914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, K.; Shimomura, T.; Hashimoto, K.; Araki, H.; Sugino, A.; Matsumoto, K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 7048–7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, M.; Okuzaki, D.; Tanaka, S.; Tougan, T.; Tamai, K.K.; Shimoda, C.; Nojima, H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol. Biol. Cell 1999, 10, 3991–4003. [Google Scholar] [CrossRef] [Green Version]
- Krause, S.A.; Loupart, M.L.; Vass, S.; Schoenfelder, S.; Harrison, S.; Heck, M.M. Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol. Cell Biol. 2001, 21, 5156–5168. [Google Scholar] [CrossRef] [Green Version]
- Britt, A.B. DNA Damage and Repair in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 75–100. [Google Scholar] [CrossRef]
- Kovalchuk, O.; Dubrova, Y.E.; Arkhipov, A.; Hohn, B.; Kovalchuk, I. Wheat mutation rate after Chernobyl. Nature 2000, 407, 583–584. [Google Scholar] [CrossRef]
- Ries, G.; Heller, W.; Puchta, H.; Sandermann, H.; Seidlitz, H.K.; Hohn, B. Elevated UV-B radiation reduces genome stability in plants. Nature 2000, 406, 98–101. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Waterworth, W.M.; Sunderland, P.A.; Bray, C.M. Arabidopsis DNA double-strand break repair pathways. Biochem. Soc. Trans. 2004, 32, 964–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, M.; Ishikawa, Y.; Osakabe, K.; Nakayama, S.; Kaya, H.; Araki, T.; Shibahara, K.; Abe, K.; Ichikawa, H.; Valentine, L.; et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006, 25, 5579–5590. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Kawashima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, J.; Miki, D.; Xia, R.; Yu, W.; He, J.; Zheng, Z.; Zhu, J.K.; Gong, Z. DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 2010, 22, 2336–2352. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Deng, Y.; Li, G.; Zhao, J. Replication factor C1 (RFC1) is required for double-strand break repair during meiotic homologous recombination in Arabidopsis. Plant J. 2013, 73, 154–165. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, Z.; Huang, J.; Shi, Q.; Hong, Y.; Copenhaver, G.P.; Gong, Z.; Ma, H. The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1003039. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Xiao, L.; Gannon, P.; Li, X. RFC3 regulates cell proliferation and pathogen resistance in Arabidopsis. Plant Signal. Behav. 2010, 5, 168–170. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Zhu, Z.; Hao, L.; Chen, J.G.; Xiao, L.; Zhang, Y.; Li, X. Negative regulation of systemic acquired resistance by replication factor C subunit3 in Arabidopsis. Plant Physiol. 2009, 150, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Acevedo-Rocha, C.G.; Reetz, M.T. Boosting the efficiency of site-saturation mutagenesis for a difficult-to-randomize gene by a two-step PCR strategy. Appl. Microbiol. Biotechnol. 2018, 102, 6095–6103. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Sterken, R.; Kiekens, R.; Boruc, J.; Zhang, F.; Vercauteren, A.; Vercauteren, I.; De Smet, L.; Dhondt, S.; Inze, D.; De Veylder, L.; et al. Combined linkage and association mapping reveals CYCD5;1 as a quantitative trait gene for endoreduplication in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 4678–4683. [Google Scholar] [CrossRef] [Green Version]
- Ohad, N.; Margossian, L.; Hsu, Y.C.; Williams, C.; Repetti, P.; Fischer, R.L. A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 1996, 93, 5319–5324. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Reidt, W.; Wurz, R.; Wanieck, K.; Chu, H.H.; Puchta, H. A homologue of the breast cancer-associated gene BARD1 is involved in DNA repair in plants. EMBO J. 2006, 25, 4326–4337. [Google Scholar] [CrossRef]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef]
- Mazin, A.V.; Mazina, O.M.; Bugreev, D.V.; Rossi, M.J. Rad54, the motor of homologous recombination. DNA Repair. (Amst) 2010, 9, 286–302. [Google Scholar] [CrossRef] [Green Version]
- Boltz, K.A.; Jasti, M.; Townley, J.M.; Shippen, D.E. Analysis of poly(ADP-Ribose) polymerases in Arabidopsis telomere biology. PLoS ONE 2014, 9, e88872. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, T.; Koga, A.; Yamamoto, T.; Uchiyama, Y.; Mori, Y.; Hashimoto, J.; Kimura, S.; Sakaguchi, K. Two types of replication protein A in seed plants. FEBS J. 2005, 272, 3270–3281. [Google Scholar] [CrossRef]
- Arbel, M.; Choudhary, K.; Tfilin, O.; Kupiec, M. PCNA Loaders and Unloaders-One Ring That Rules Them All. Genes (Basel) 2021, 12, 1812. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Maintenance of genome stability in plants: Repairing DNA double strand breaks and chromatin structure stability. Front. Plant Sci. 2014, 5, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Choudhury, S.R.; Sengupta, D.N.; Das, K.P. Involvement of AtPolλ in the repair of high salt- and DNA cross-linking agent-induced double strand breaks in Arabidopsis. Plant Physiol. 2013, 162, 1195–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [Green Version]
- Heitzeberg, F.; Chen, I.P.; Hartung, F.; Orel, N.; Angelis, K.J.; Puchta, H. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 2004, 38, 954–968. [Google Scholar] [CrossRef]
- Lundin, C.; North, M.; Erixon, K.; Walters, K.; Jenssen, D.; Goldman, A.S.; Helleday, T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005, 33, 3799–3811. [Google Scholar] [CrossRef]
- Schuermann, D.; Fritsch, O.; Lucht, J.M.; Hohn, B. Replication stress leads to genome instabilities in Arabidopsis DNA polymerase delta mutants. Plant Cell 2009, 21, 2700–2714. [Google Scholar] [CrossRef] [Green Version]
- Shaul, O.; Mironov, V.; Burssens, S.; Van Montagu, M.; Inze, D. Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 1996, 93, 4868–4872. [Google Scholar] [CrossRef] [Green Version]
- Yi, D.; Alvim Kamei, C.L.; Cools, T.; Vanderauwera, S.; Takahashi, N.; Okushima, Y.; Eekhout, T.; Yoshiyama, K.O.; Larkin, J.; Van den Daele, H.; et al. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 2014, 26, 296–309. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, N.; Hernould, M.; Delmas, F.; Gevaudant, F.; Duffe, P.; Causse, M.; Mouras, A.; Chevalier, C. Molecular characterization of a WEE1 gene homologue in tomato (Lycopersicon esculentum Mill.). Plant Mol. Biol. 2004, 56, 849–861. [Google Scholar] [CrossRef]
- Gray, F.C.; MacNeill, S.A. The Schizosaccharomyces pombe rfc3+ gene encodes a homologue of the human hRFC36 and Saccharomyces cerevisiae Rfc3 subunits of replication factor C. Curr. Genet. 2000, 37, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.; Fantes, P.A.; MacNeill, S.A. A key role for replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 1999, 27, 462–469. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, K.; Qin, L.; Tang, X.; Nong, J.; Chen, J.; Wu, N.; Gong, X.; Yi, L.; Yang, C.; Xia, S. A Single Amino Acid Substitution in RFC4 Leads to Endoduplication and Compromised Resistance to DNA Damage in Arabidopsis thaliana. Genes 2022, 13, 1037. https://doi.org/10.3390/genes13061037
Cui K, Qin L, Tang X, Nong J, Chen J, Wu N, Gong X, Yi L, Yang C, Xia S. A Single Amino Acid Substitution in RFC4 Leads to Endoduplication and Compromised Resistance to DNA Damage in Arabidopsis thaliana. Genes. 2022; 13(6):1037. https://doi.org/10.3390/genes13061037
Chicago/Turabian StyleCui, Kan, Lei Qin, Xianyu Tang, Jieying Nong, Jin Chen, Nan Wu, Xin Gong, Lixiong Yi, Chenghuizi Yang, and Shitou Xia. 2022. "A Single Amino Acid Substitution in RFC4 Leads to Endoduplication and Compromised Resistance to DNA Damage in Arabidopsis thaliana" Genes 13, no. 6: 1037. https://doi.org/10.3390/genes13061037
APA StyleCui, K., Qin, L., Tang, X., Nong, J., Chen, J., Wu, N., Gong, X., Yi, L., Yang, C., & Xia, S. (2022). A Single Amino Acid Substitution in RFC4 Leads to Endoduplication and Compromised Resistance to DNA Damage in Arabidopsis thaliana. Genes, 13(6), 1037. https://doi.org/10.3390/genes13061037