Epigenetic Modulations for Prevention of Infectious Diseases in Shrimp Aquaculture
Abstract
:1. Introduction
2. Current Status of Global Production and Challenges in Shrimp Aquaculture


3. Overview of Current Shrimp Disease Management Strategies
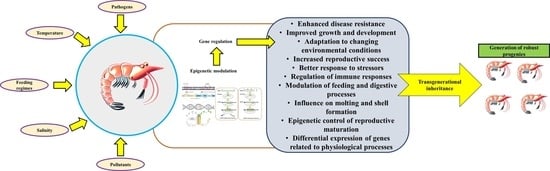
4. The Concept of Epigenetics, Epigenetic Inheritance, and Its Potential Effect on Disease Management in Shrimp
5. Types of Epigenetic Modifications
5.1. DNA Methylation
5.2. Histone Modifications

5.3. Non-Coding RNA

6. Applications of Epigenetics for Immunity Enhancement and Disease Management in Shrimp Aquaculture
7. Challenges and Future Perspectives of Epigenetics in the Disease Management of Shrimp Aquaculture
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO Publications Catalogue 2022; FAO: Rome, Italy, 2022. [CrossRef]
- Araujo, G.S.; da Silva, J.W.A.; Cotas, J.; Pereira, L. Fish farming techniques: Current situation and trends. J. Mar. Sci. Eng. 2022, 10, 1598. [Google Scholar] [CrossRef]
- Ahmed, N.; Azra, M.N. Aquaculture production and value chains in the COVID-19 pandemic. Curr. Environ. Health Rep. 2022, 9, 423–435. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture. 2020. pp. 1–244. Available online: http://www.fao.org/3/ca9229en/ca9229en.pdf (accessed on 18 August 2023).
- Gunalan, B.; Nina Tabitha, S.; Soundarapandian, P.; Anand, T. Nutritive value of cultured white leg shrimp Litopenaeus vannamei. Int. J. Fish. Aquac. 2013, 5, 166–171. [Google Scholar]
- Shrimps—A Nutritional Perspective on JSTOR. Available online: https://www.jstor.org/stable/24092471 (accessed on 21 July 2023).
- Kulkarni, A.; Krishnan, S.; Anand, D.; Kokkattunivarthil Uthaman, S.; Otta, S.K.; Karunasagar, I.; Kooloth Valappil, R. Immune responses and immunoprotection in crustaceans with special reference to shrimp. Rev. Aquac. 2021, 13, 431–459. [Google Scholar] [CrossRef]
- Millard, R.S.; Ellis, R.P.; Bateman, K.S.; Bickley, L.K.; Tyler, C.R.; van Aerle, R.; Santos, E.M. How do abiotic environmental conditions influence shrimp susceptibility to disease? A critical analysis focussed on white spot disease. J. Invertebr. Pathol. 2021, 186, 107369. [Google Scholar] [CrossRef]
- Bank, T.W. Reducing disease risk in aquaculture. World Bank. Agric. Environ. Serv. 2014, 119, 1–6. [Google Scholar]
- Yu, Y.B.; Choi, J.H.; Kang, J.C.; Kim, H.J.; Kim, J.H. Shrimp bacterial and parasitic disease listed in the OIE: A review. Microb. Pathog. 2022, 166, 105545. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Jiang, Y. The use of chemicals in aquaculture in the people’s Republic of China. In Proceedings of the Meeting on the Use of Chemicals in Aquaculture in Asia, Iloilo, Philippines, 20–22 May 1996; pp. 141–153. [Google Scholar]
- Li, C.; Lin, N.; Feng, Z.; Lin, M.; Guan, B.; Chen, K.; Liang, W.; Wang, Q.; Li, M.; You, Y.; et al. CRISPR/Cas12a based rapid molecular detection of acute hepatopancreatic necrosis disease in shrimp. Front. Vet. Sci. 2022, 8, 819681. [Google Scholar] [CrossRef]
- Sivakamavalli, J.; Park, K.; Kwak, I.; Baskaralingam, V. Bacterial disease control methods in shrimp (Penaeus, 1798) farming sector in Asian countries. In Arthropods are They Benef. Mankind; Intechopen: London, UK, 2021. [Google Scholar] [CrossRef]
- Roy, S.; Kumar, V.; Bossier, P.; Norouzitallab, P.; Vanrompay, D. Phloroglucinol treatment induces transgenerational epigenetic inherited resistance against vibrio infections and thermal stress in a brine shrimp (Artemia franciscana) model. Front. Immunol. 2019, 10, 2745. [Google Scholar] [CrossRef]
- Granada, L.; Lemos, M.F.L.; Cabral, H.N.; Bossier, P.; Novais, S.C. Epigenetics in aquaculture—The last frontier. Rev. Aquac. 2018, 10, 994–1013. [Google Scholar] [CrossRef]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenet. 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xue, Y. Emerging roles of non-coding RNAs in epigenetic regulation. Sci. China Life Sci. 2016, 59, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Daxinger, L.; Whitelaw, E. Transgenerational epigenetic inheritance: More questions than answers. Genome Res. 2010, 20, 1623–1628. [Google Scholar] [CrossRef]
- Lacal, I.; Ventura, R. Epigenetic inheritance: Concepts, mechanisms and perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef]
- Aguilera, O.; Fernández, A.F.; Muñoz, A.; Fraga, M.F. Epigenetics and environment: A complex relationship. J. Appl. Physiol. 2010, 109, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Baruah, K.; Vandegehuchte, M.; Van Stappen, G.; Catania, F.; Vanden Bussche, J.; Vanhaecke, L.; Sorgeloos, P.; Bossier, P. Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic artemia model. FASEB J. 2014, 28, 3552–3563. [Google Scholar] [CrossRef]
- Manan, H.; Ikhwanuddin, M. Triploid induction in penaeid shrimps aquaculture: A review. Rev. Aquac. 2021, 13, 619–631. [Google Scholar] [CrossRef]
- IMARC Group. Shrimp Market: Global Industry Trends, Share, Size, Growth, Opportunity and Forecast 2020–2025. 2020. Available online: https://www.imarcgroup.com/prefeasibility-report-shrimp-processing-plant (accessed on 18 August 2023).
- Tabarestani, M.; Keithly, W.R.; Marzoughi-Ardakani, H. An analysis of the US shrimp market: A mixed demand approach. Mar. Resour. Econ. 2017, 32, 411–429. [Google Scholar] [CrossRef]
- Jory; Darryl, E. Current production, challenges and the future of shrimp farming. Glob. Aquac. Alliance 2018, 1, 1–8. [Google Scholar]
- Suzuki, A.; Nam, V.H. Blue Revolution in Asia: The Rise of the Shrimp Sector in Vietnam and the Challenges of Disease Control; Springer: Berlin, Germany, 2023; pp. 289–303. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Swelum, A.A.; Abo Ghanima, M.M.; Shukry, M.; Omar, A.A.; Taha, A.E.; Salem, H.M.; El-Tahan, A.M.; El-Tarabily, K.A.; Abd El-Hack, M.E. Shrimp production, the most important diseases that threaten it, and the role of probiotics in confronting these diseases: A review. Res. Vet. Sci. 2022, 144, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wang, X.; Chen, K.; Xu, C.; Qin, J.G.; Chen, L. Physiological change and nutritional requirement of pacific white shrimp Litopenaeus vannamei at low salinity. Rev. Aquac. 2017, 9, 57–75. [Google Scholar] [CrossRef]
- Li, E.; Xu, C.; Wang, X.; Wang, S.; Zhao, Q.; Zhang, M.; Qin, J.G.; Chen, L. Gut microbiota and its modulation for healthy farming of pacific white shrimp Litopenaeus vannamei. Rev. Fish. Sci. Aquac. 2018, 26, 381–399. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.H.; Pinto, A.R. Challenges in shrimp aquaculture due to viral diseases: Distribution and biology of the five major penaeid viruses and interventions to avoid viral incidence and dispersion. Brazilian J. Microbiol. 2012, 43, 857–864. [Google Scholar] [CrossRef]
- Lee, D.; Yu, Y.B.; Choi, J.H.; Jo, A.H.; Hong, S.M.; Kang, J.C.; Kim, J.H. Viral shrimp diseases listed by the OIE: A review. Viruses 2022, 14, 585. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Poulos, B.T.; Nunan, L.M.; Mari, J.L.; Hasson, K.W. Risk of spread of penaeid shrimp viruses in the Americas by the international movement of live and frozen shrimp. OIE Rev. Sci. Tech. 1997, 16, 146–160. [Google Scholar] [CrossRef]
- APEC. Trans-Boundary Aquatic Animal Pathogen Transfer and the Development of Harmonized Standards on Aquaculture Health; Erawan Interactive: Puerto Vallarta, Mexico, 2001; ISBN 9747313278. [Google Scholar]
- Jones, B. Transboundary movement of shrimp viruses in crustaceans and their products: A special risk? J. Invertebr. Pathol. 2012, 110, 196–200. [Google Scholar] [CrossRef]
- Karunasagar, I.; Ababouch, L. Shrimp viral diseases, import risk assessment and international trade. Indian J. Virol. 2012, 23, 141–148. [Google Scholar] [CrossRef]
- Flegel, T.W.; Alday-Sanz, V. The crisis in Asian shrimp aquaculture: Current status and future needs. J. Appl. Ichthyol. 1998, 14, 269–273. [Google Scholar] [CrossRef]
- Stentiford, G.D.; Neil, D.M.; Peeler, E.J.; Shields, J.D.; Small, H.J.; Flegel, T.W.; Vlak, J.M.; Jones, B.; Morado, F.; Moss, S.; et al. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012, 110, 141–157. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Pantoja, C.R.; Poulos, B.T.; Redman, R.M.; Lightner, D.V. In situ hybridization demonstrates that Litopenaeus vannamei, L. stylirostris and Penaeus monodon are susceptible to experimental infection with infectious myonecrosis virus (IMNV). Dis. Aquat. Organ. 2005, 63, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Andrade, T.P.D.; Cruz-Flores, R.; Mai, H.N.; Dhar, A.K. Novel infectious myonecrosis virus (IMNV) variant is associated with recent disease outbreaks in Penaeus vannamei shrimp in Brazil. Aquaculture 2022, 554, 738159. [Google Scholar] [CrossRef]
- Feijó, R.G.; Kamimura, M.T.; Oliveira-Neto, J.M.; Vila-Nova, C.M.V.M.; Gomes, A.C.S.; Coelho, M.d.G.L.; Vasconcelos, R.F.; Gesteira, T.C.V.; Marins, L.F.; Maggioni, R. Infectious myonecrosis virus and white spot syndrome virus co-infection in pacific white shrimp (Litopenaeus vannamei) farmed in Brazil. Aquaculture 2013, 380–383, 1–5. [Google Scholar] [CrossRef]
- The State of World Fisheries and Aquaculture; FAO: Rome, Italy, 2022. [CrossRef]
- Gomez-Gil, B.; Soto-Rodríguez, S.; Lozano, R.; Betancourt-Lozano, M. Draft genome sequence of Vibrio parahaemolyticus strain m0605, which causes severe mortalities of shrimps in Mexico. Genome Announc. 2014, 2, 2104. [Google Scholar] [CrossRef]
- Lightner, D.V.; Ph, D. Early mortality syndrome affects shrimp in Asia. Glob. Aquac. Advocate 2012, 40, 2012. [Google Scholar]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ. 2013, 105, 45–55. [Google Scholar] [CrossRef]
- FAO. Report of the FAO/MARD Technical Workshop on Early Mortality Syndrome (EMS) or Acute Hepatopancreatic Necrosis Syndrome (AHPNS) of Cultured Shrimp (under TCP/VIE/3304); FAO: Hanoi, Vietnam, 2013; Volume 1053, ISBN 9789251079041. [Google Scholar]
- Aranguren Caro, L.F.; Mai, H.N.; Noble, B.; Dhar, A.K. Acute hepatopancreatic necrosis disease (VpAHPND), a chronic disease in shrimp (Penaeus vannamei) population raised in Latin America. J. Invertebr. Pathol. 2020, 174, 107424. [Google Scholar] [CrossRef]
- Hossain, M.S.; Dai, J.; Qiu, D. Dysbiosis of the shrimp (Penaeus monodon) gut microbiome with AHPND outbreaks revealed by 16S rRNA metagenomics analysis. Aquac. Res. 2021, 52, 3336–3349. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Kumar, P.; Sivachandran, P.; Marimuthu, K.; Arshad, A.; Paray, B.A.; Arockiaraj, J. Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 2020, 12, 1903–1927. [Google Scholar] [CrossRef]
- Rahi, M.L.; Azad, K.N.; Tabassum, M.; Irin, H.H.; Hossain, K.S.; Aziz, D.; Moshtaghi, A.; Hurwood, D.A. Effects of salinity on physiological, biochemical and gene expression parameters of black tiger shrimp (Penaeus monodon): Potential for farming in low-salinity environments. Biology 2021, 10, 1220. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wang, Q.; Shao, H.; Xu, Y.; Liu, P.; Li, J. Effects of low temperature on shrimp and crab physiology, behavior, and growth: A review. Front. Mar. Sci. 2021, 8, 746177. [Google Scholar] [CrossRef]
- Kataoka, C.; Kashiwada, S. Ecological risks due to immunotoxicological effects on aquatic organisms. Int. J. Mol. Sci. 2021, 22, 8305. [Google Scholar] [CrossRef]
- Argue, B.B.J.; Tolentino, G.; Brock, J.A. Inbreeding Cuts Growth, Reproduction in Shrimp—Responsible Seafood Advocate. 6 April 2018; pp. 7–11. Available online: https://www.globalseafood.org/advocate/inbreeding-cuts-growth-reproduction-in-shrimp/ (accessed on 18 August 2023).
- Doyle, R.W. Inbreeding and disease in tropical shrimp aquaculture: A reappraisal and caution. Aquac. Res. 2016, 47, 21–35. [Google Scholar] [CrossRef]
- Asche, F.; Anderson, J.L.; Botta, R.; Kumar, G.; Abrahamsen, E.B.; Nguyen, L.T.; Valderrama, D. The economics of shrimp disease. J. Invertebr. Pathol. 2021, 186, 107397. [Google Scholar] [CrossRef] [PubMed]
- An Update on Vibriosis, the Major Bacterial Disease Shrimp Farmers Face—Responsible Seafood Advocate. Available online: https://www.globalseafood.org/advocate/an-update-on-vibriosis-the-major-bacterial-disease-shrimp-farmers-face/ (accessed on 18 August 2023).
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Davies, J. Origins and evolution of antibiotic resistance. Microbiologia 1996, 12, 9–16. [Google Scholar] [CrossRef]
- Davis, R.P.; Davis, D.A.; Boyd, C.E. A preliminary survey of antibiotic residues in frozen shrimp from retail stores in the United States. Curr. Res. Food Sci. 2021, 4, 679–683. [Google Scholar] [CrossRef]
- Le, T.X.; Munekage, Y. Residues of selected antibiotics in water and mud from shrimp ponds in mangrove areas in Viet Nam. Mar. Pollut. Bull. 2004, 49, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Nagpal, R.; Jackson, C.R.; Patel, D.; Singh, P. Antibiotic-resistant bacteria and gut microbiome communities associated with wild-caught shrimp from the United States versus imported farm-raised retail shrimp. Sci. Rep. 2021, 11, 3356. [Google Scholar] [CrossRef]
- Thornber, K.; Verner-Jeffreys, D.; Hinchliffe, S.; Rahman, M.M.; Bass, D.; Tyler, C.R. Evaluating antimicrobial resistance in the global shrimp industry. Rev. Aquac. 2020, 12, 966–986. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, J.; Zhao, Z.; Chen, J.; Lu, H.; Liu, G. Fishmeal application induces antibiotic resistance gene propagation in mariculture sediment. Environ. Sci. Technol. 2017, 51, 10850–10860. [Google Scholar] [CrossRef] [PubMed]
- Sudheesh, P.S.; Cain, K.D. Prospects and challenges of developing and commercializing immersion vaccines for aquaculture. Int. Biol. Rev. 2017, 1, 1–20. [Google Scholar]
- Gudding, R.; Van Muiswinkel, W.B. A history of fish vaccination: Science-based disease prevention in aquaculture. Fish Shellfish Immunol. 2013, 35, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Bruce, T.J.; Jones, E.M.; Cain, K.D. A Review of fish vaccine development strategies: Conventional methods and modern biotechnological approaches. Microorganisms 2019, 7, 569. [Google Scholar] [CrossRef] [PubMed]
- Rout, N.; Kumar, S.; Jaganmohan, S.; Murugan, V. DNA vaccines encoding viral envelope proteins confer protective immunity against WSSV in black tiger shrimp. Vaccine 2007, 25, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Rajeshkumar, S.; Venkatesan, C.; Sarathi, M.; Sarathbabu, V.; Thomas, J.; Anver Basha, K.; Sahul Hameed, A.S. Oral delivery of DNA construct using chitosan nanoparticles to protect the shrimp from white spot syndrome virus (WSSV). Fish Shellfish Immunol. 2009, 26, 429–437. [Google Scholar] [CrossRef]
- Mavichak, R.; Takano, T.; Kondo, H.; Hirono, I.; Wada, S.; Hatai, K.; Inagawa, H.; Takahashi, Y.; Yoshimura, T.; Kiyono, H.; et al. The effect of liposome-coated recombinant protein VP28 against white spot syndrome virus in kuruma shrimp, Marsupenaeus japonicus. J. Fish Dis. 2010, 33, 69–74. [Google Scholar] [CrossRef]
- Patil, P.K.; Gopal, C.; Panigrahi, A.; Rajababu, D.; Pillai, S.M. Oral administration of formalin killed Vibrio anguillarum cells improves growth and protection against challenge with Vibrio harveyi in banana shrimp. Lett. Appl. Microbiol. 2014, 58, 213–218. [Google Scholar] [CrossRef]
- Rowley, A.F.; Pope, E.C. Vaccines and crustacean aquaculture—A mechanistic exploration. Aquaculture 2012, 334–337, 1–11. [Google Scholar] [CrossRef]
- Ramos-Vivas, J.; Superio, J.; Galindo-Villegas, J.; Acosta, F. Phage therapy as a focused management strategy in aquaculture. Int. J. Mol. Sci. 2021, 22, 10436. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Sillankorva, S.M.; Oliveira, H.; Azeredo, J. Bacteriophages and their role in food safety. Int. J. Microbiol. 2012, 2012. [Google Scholar] [CrossRef]
- Jun, J.W.; Han, J.E.; Giri, S.S.; Tang, K.F.J.; Zhou, X.; Aranguren, L.F.; Kim, H.J.; Yun, S.; Chi, C.; Kim, S.G.; et al. Phage application for the protection from acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei. Indian J. Microbiol. 2018, 58, 114–117. [Google Scholar] [CrossRef]
- Quiroz-Guzmán, E.; Peña-Rodriguez, A.; Vázquez-Juárez, R.; Barajas-Sandoval, D.R.; Balcázar, J.L.; Martínez-Díaz, S.F. Bacteriophage cocktails as an environmentally-friendly approach to prevent Vibrio parahaemolyticus and Vibrio harveyi infections in brine shrimp (Artemia franciscana) production. Aquaculture 2018, 492, 273–279. [Google Scholar] [CrossRef]
- Yin, Y.; Ni, P.; Liu, D.; Yang, S.; Almeida, A.; Guo, Q.; Zhang, Z.; Deng, L.; Wang, D. Bacteriophage potential against Vibrio parahaemolyticus biofilms. Food Control 2019, 98, 156–163. [Google Scholar] [CrossRef]
- Borodovich, T.; Shkoporov, A.N.; Ross, R.P.; Hill, C. Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol. Rep. 2022, 10, goac012. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Skurnik, M.; Strauch, E. Phage therapy: Facts and fiction. Int. J. Med. Microbiol. 2006, 296, 5–14. [Google Scholar] [CrossRef]
- Sakai, M. Current research status of fish immunostimulants. Aquaculture 1999, 172, 63–92. [Google Scholar] [CrossRef]
- Declarador, R.S.; Serrano, A.E.; Corre, V.L. Ulvan extract acts as immunostimulant against white spot syndrome virus (WSSV) in juvenile black tiger shrimp Penaeus monodon. AACL Bioflux 2014, 7, 153–161. [Google Scholar]
- Citarasu, T. Herbal biomedicines: A new opportunity for aquaculture industry. Aquac. Int. 2010, 18, 403–414. [Google Scholar] [CrossRef]
- Anirudhan, A.; Tosin, O.V.; Wahid, M.; Effendy; Sung, Y.Y. Alternative disease control methods in shrimp aquaculture: A review. J. Aquac. Res. Dev. 2021, 12, 12–15. [Google Scholar]
- Yu, P.; Wang, T.; Ye, H.; Shan, H.; Ma, S. Isolation and identification of pathogenic vibrio spp. retrieved from diseased Litopenaeus vannamei and beneficial role of some functional probiotic bacteria for control. Aquac. Int. 2020, 28, 1403–1420. [Google Scholar] [CrossRef]
- Khademzade, O.; Zakeri, M.; Haghi, M.; Mousavi, S.M. The effects of water additive Bacillus cereus and Pediococcus acidilactici on water quality, growth performances, economic benefits, immunohematology and bacterial flora of whiteleg shrimp (Penaeus vannamei boone, 1931) reared in earthen ponds. Aquac. Res. 2020, 51, 1759–1770. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Dai, X.; Li, J.; Ding, F. Effects of dietary inulin and mannan oligosaccharide on immune related genes expression and disease resistance of pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2018, 76, 78–92. [Google Scholar] [CrossRef]
- Rungrassamee, W.; Kingcha, Y.; Srimarut, Y.; Maibunkaew, S.; Karoonuthaisiri, N.; Visessanguan, W. Mannooligosaccharides from copra meal improves survival of the pacific white shrimp (Litopenaeus vannamei) after exposure to Vibrio harveyi. Aquaculture 2014, 434, 403–410. [Google Scholar] [CrossRef]
- Huynh, T.G.; Cheng, A.C.; Chi, C.C.; Chiu, K.H.; Liu, C.H. A synbiotic improves the immunity of white shrimp, Litopenaeus vannamei: Metabolomic analysis reveal compelling evidence. Fish Shellfish Immunol. 2018, 79, 284–293. [Google Scholar] [CrossRef]
- Roy, S.; Kumar, V.; Behera, B.K.; Das, B.K. Epigenetics: Perspectives and potential in aquaculture. Adv. Fish. Biotechnol. 2022, 133–150. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, T.; Gao, D. Genetic and epigenetic regulation of growth, reproduction, disease resistance and stress responses in aquaculture. Front. Genet. 2022, 13, 994471. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J. The changing concept of epigenetics. Ann. N. Y. Acad. Sci. 2002, 981, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Brander, S.M.; Biales, A.D.; Connon, R.E. The role of epigenomics in aquatic toxicology. Environ. Toxicol. Chem. 2017, 36, 2565–2573. [Google Scholar] [CrossRef]
- Skinner, M.K. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res. C Embryo Today 2011, 93, 51. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Yang, J.J.; Shi, K.H. Non-Coding RNAs as direct and indirect modulators of epigenetic mechanism regulation of cardiac fibrosis. Expert Opin. Ther. Targets 2015, 19, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-Coding RNAs as regulators in epigenetics (review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Biswas, P.; Baruah, K.; Bossier, P. Multigenerational immune priming in an invertebrate parthenogenetic artemia to a pathogenic Vibrio campbellii. Fish Shellfish Immunol. 2015, 42, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95. [Google Scholar] [CrossRef]
- Norouzitallab, P.; Baruah, K.; Biswas, P.; Vanrompay, D.; Bossier, P. Probing the phenomenon of trained immunity in invertebrates during a transgenerational study, using brine shrimp artemia as a model system. Sci. Rep. 2016, 6, 21166. [Google Scholar] [CrossRef]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Miremadi, A.; Oestergaard, M.Z.; Pharoah, P.D.P.; Caldas, C. Cancer genetics of epigenetic genes. Human Mol. Genet. 2007, 16, R28–R49. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA ( cytosine-5 ) methyltransferases non-invasive sexing of preimplantation stage mammalian embryos. Nat. Am. Inc. 1998, 19, 219–220. [Google Scholar]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Engelhardt, J.; Scheer, O.; Stadler, P.F.; Prohaska, S.J. Evolution of DNA methylation across ecdysozoa. J. Mol. Evol. 2022, 90, 56–72. [Google Scholar] [CrossRef]
- Lee, H.J.; Lowdon, R.F.; Maricque, B.; Zhang, B.; Stevens, M.; Li, D.; Johnson, S.L.; Wang, T. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat. Commun. 2015, 6, 6315. [Google Scholar] [CrossRef]
- Rauscher, G.H.; Kresovich, J.K.; Poulin, M.; Yan, L.; Macias, V.; Mahmoud, A.M.; Al-Alem, U.; Kajdacsy-Balla, A.; Wiley, E.L.; Tonetti, D.; et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer 2015, 15, 1–15. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, J.; Romanovskaia, D.; Nemc, A.; Posautz, A.; Seid, C.A.; Schuster, L.C.; Keinath, M.C.; Lugo Ramos, J.S.; Kosack, L.; Evankow, A.; et al. Comparative analysis of genome-scale, base-resolution DNA methylation profiles across 580 animal species. Nat. Commun. 2023, 14, 232. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Hon, G.C.; Rajagopal, N.; Shen, Y.; McCleary, D.F.; Yue, F.; Dang, M.D.; Ren, B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013, 45, 1198–1206. [Google Scholar] [CrossRef]
- Marhold, J.; Kramer, K.; Kremmer, E.; Lyko, F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development 2004, 131, 6033–6039. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Han, H.; DeCarvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Shayevitch, R.; Askayo, D.; Keydar, I.; Ast, G. The importance of DNA methylation of exons on alternative splicing. Rna 2018, 24, 1351–1362. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Sukthaworn, S.; Panyim, S.; Udomkit, A. Homologues of piwi control transposable elements and development of male germline in Penaeus monodon. Comp. Biochem. Physiol.-Part A Mol. Integr. Physiol. 2020, 250, 110807. [Google Scholar] [CrossRef]
- Li, B.B.; Fan, J.Q.; Lu, K.C.; Chen, G.L.; Chen, Y.H. Identification and functional characterization of a systemic RNA interference defective 1 gene in Litopenaeus vannamei. Fish Shellfish Immunol. Rep. 2021, 2, 100033. [Google Scholar] [CrossRef]
- Shilatifard, A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006, 75, 243–269. [Google Scholar] [CrossRef]
- Marshall, O.J.; Brand, A.H. Chromatin state changes during neural development revealed by in vivo cell-type specific profiling. Nat. Commun. 2017, 8, 2271. [Google Scholar] [CrossRef]
- Henikoff, S.; Shilatifard, A. Histone modification: Cause or cog? Trends Genet. 2011, 27, 389–396. [Google Scholar] [CrossRef]
- Turner, B.M.; Birley, A.J.; Lavender, J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 1992, 69, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 421, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Duman, M.; Martinez-Moreno, M.; Jacob, C.; Tapinos, N. Functions of histone modifications and histone modifiers in schwann cells. Glia 2020, 68, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, Y. Mapping post-translational modifications of histones H2A, H2B and H4 in Schizosaccharomyces pombe. Int. J. Mass Spectrom. 2011, 301, 159. [Google Scholar] [CrossRef]
- Karmodiya, K.; Anamika, K.; Muley, V.; Pradhan, S.J.; Bhide, Y.; Galande, S. Camello, A novel family of histone acetyltransferases that acetylate histone H4 and is essential for zebrafish development. Sci. Rep. 2014, 4, 6076. [Google Scholar] [CrossRef]
- Yang, J.; Yang, S.; Liao, Y.; Deng, Y.; Jiao, Y. Increased histone H3 acetylation inhibit the inflammatory response and activate the serum immunity of pearl oyster Pinctada fucata martensii. Front. Mar. Sci. 2023, 10, 1073322. [Google Scholar] [CrossRef]
- Chatterjee, N.; Sinha, D.; Lemma-Dechassa, M.; Tan, S.; Shogren-Knaak, M.A.; Bartholomew, B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011, 39, 8378–8391. [Google Scholar] [CrossRef]
- Wakamori, M.; Okabe, K.; Ura, K.; Funatsu, T.; Takinoue, M.; Umehara, T. Quantification of the effect of site-specific histone acetylation on chromatin transcription rate. Nucleic Acids Res. 2020, 48, 12648–12659. [Google Scholar] [CrossRef]
- Patat, S.A.; Carnegie, R.B.; Kingsbury, C.; Gross, P.S.; Chapman, R.; Schey, K.L. Antimicrobial activity of histones from hemocytes of the pacific white shrimp. Eur. J. Biochem. 2004, 271, 4825–4833. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Lauberth, S.M.; Nakayama, T.; Wu, X.; Ferris, A.L.; Tang, Z.; Hughes, S.H.; Roeder, R.G. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013, 152, 1021–1036. [Google Scholar] [CrossRef] [PubMed]
- Gillies, S.D.; Morrison, S.L.; Oi, V.T.; Tonegawa, S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 1983, 33, 717–728. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Brickner, J.H. Epigenetic transcriptional memory. Curr. Genet. 2017, 63, 435. [Google Scholar] [CrossRef]
- Zang, S.; Lv, L.X.; Liu, C.F.; Zhang, P.; Li, C.; Wang, J.X. Metabolomic investigation of ultraviolet ray-inactivated white spot syndrome virus-induced trained immunity in Marsupenaeus japonicus. Front. Immunol. 2022, 13, 885782. [Google Scholar] [CrossRef] [PubMed]
- Baarends, W.M.; Hoogerbrugge, J.W.; Roest, H.P.; Ooms, M.; Vreeburg, J.; Hoeijmakers, J.H.J.; Grootegoed, J.A. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 1999, 207, 322–333. [Google Scholar] [CrossRef]
- Boros, I.M. Histone modification in Drosophila. Brief. Funct. Genom. 2012, 11, 319–331. [Google Scholar] [CrossRef]
- Gonzalez-Romero, R.; Suarez-Ulloa, V.; Rodriguez-Casariego, J.; Garcia-Souto, D.; Diaz, G.; Smith, A.; Pasantes, J.J.; Rand, G.; Eirin-Lopez, J.M. Effects of florida red tides on histone variant expression and DNA methylation in the eastern oyster Crassostrea virginica. Aquat. Toxicol. 2017, 186, 196–204. [Google Scholar] [CrossRef]
- Teperek-Tkacz, M.; Meglicki, M.; Pasternak, M.; Kubiak, J.Z.; Borsuk, E. Phosphorylation of histone H3 serine 10 in early mouse embryos: Active phosphorylation at late s phase and differential effects of ZM447439 on first two embryonic mitoses. Cell Cycle 2010, 9, 4674–4687. [Google Scholar] [CrossRef]
- Kellner, W.A.; Ramos, E.; Van Bortle, K.; Takenaka, N.; Corces, V.G. Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters. Genome Res. 2012, 22, 1081–1088. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Jin, Y.; Johansen, J.; Johansen, K.M. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 2001, 105, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Song, Z.; Li, G.; Tu, H.; Liu, W.; Liu, Y.; Wang, P.; Wang, Y.; Cui, X.; Liu, C.; et al. H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. Nucleic Acids Res. 2016, 44, 9681–9697. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Buszczak, M.; Paterno, S.; Spradling, A.C. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 2009, 323, 248. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Sun, Z.; Mu, S.; Jiang, L.; Li, C.; Li, L.; Guo, M.; Zhang, Z.; Kang, X. Ultrastructure of spermiogenesis and the distribution of spermatozoal nuclear histones in the Japanese mantis shrimp, Oratosquilla oratoria (Crustacea: Stomatopoda). J. Morphol. 2019, 280, 1170–1184. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Mu, S.M.; Guo, M.S.; Wu, J.L.; Li, Y.Q.; Zhang, H.; Wang, Y.; Kang, X.J. Dynamics of histone H2A, H4 and HS1ph during spermatogenesis with a focus on chromatin condensation and maturity of spermatozoa. Sci. Rep. 2016, 6, 25089. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, R.; Aweya, J.J.; Yao, D.; Wang, F.; Li, S.; Tuan, T.N.; Zhang, Y. The PirB toxin protein from Vibrio parahaemolyticus induces apoptosis in hemocytes of Penaeus vannamei: PirB induces shrimp hemocytes apoptosis. Virulence 2021, 12, 481–492. [Google Scholar] [CrossRef]
- Tang, X.; Liu, T.; Li, X.; Sheng, X.; Xing, J.; Chi, H.; Zhan, W. Protein phosphorylation in hemocytes of Fenneropenaeus chinensis in response to white spot syndrome virus infection. Fish Shellfish Immunol. 2022, 122, 106–114. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution—trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef]
- Zhou, Z.; Leng, C.; Wang, Z.; Long, L.; Lv, Y.; Gao, Z.; Wang, Y.; Wang, S.; Li, P. The potential regulatory role of the LncRNA-MiRNA-MRNA axis in teleost fish. Front. Immunol. 2023, 14, 1065357. [Google Scholar] [CrossRef] [PubMed]
- Cecere, G. Small RNAs in epigenetic inheritance: From mechanisms to trait transmission. FEBS Lett. 2021, 595, 2953–2977. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.Y.; Chang, D.C.; Lin, S.L. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008, 38, 257–268. [Google Scholar] [CrossRef]
- Brown, W.; Bardhan, A.; Darrah, K.; Tsang, M.; Deiters, A. Optical control of microRNA function in zebrafish embryos. J. Am. Chem. Soc. 2022, 144, 16819–16826. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, Y.; Guo, M.; Guo, M.; Yue, D.; Yue, D.; Chen, C.; Chen, C.; Liang, G.; Liang, G.; et al. MicroRNA-7: Expression and function in brain physiological and pathological processes. Cell Biosci. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 1–19. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, Z.; Wang, H.; Wang, L.; Wang, W.; Liu, R.; Qiu, L.; Song, L. An invertebrate-specific and immune-responsive microRNA augments oyster hemocyte phagocytosis by targeting CgIκ B2. Sci. Rep. 2016, 6, 29591. [Google Scholar] [CrossRef]
- Ruan, L.; Bian, X.; Ji, Y.; Li, M.; Li, F.; Yan, X. Isolation and identification of novel microRNAs from Marsupenaeus japonicus. Fish Shellfish Immunol. 2011, 31, 334–340. [Google Scholar] [CrossRef]
- Kaewkascholkul, N.; Somboonviwat, K.; Asakawa, S.; Hirono, I.; Tassanakajon, A.; Somboonwiwat, K. Shrimp MiRNAs regulate innate immune response against white spot syndrome virus infection. Dev. Comp. Immunol. 2016, 60, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Wang, Z.; Sun, B.Z. Differential expression of microRNAs in shrimp Marsupenaeus japonicus in response to Vibrio alginolyticus infection. Dev. Comp. Immunol. 2016, 55, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Aweya, J.J.; Wang, F.; Yao, D.; Lun, J.; Li, S.; Ma, H.; Zhang, Y. Acute hepatopancreatic necrosis disease (AHPND) related microRNAs in Litopenaeus vannamei infected with AHPND-causing strain of Vibrio parahemolyticus. BMC Genom. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Zheng, J.; Cao, J.; Mao, Y.; Su, Y.; Wang, J. Identification of microRNAs with heat stress responsive and immune properties in Marsupenaeus japonicus based on next-generation sequencing and bioinformatics analysis: Essential regulators in the heat stress-host interactions. Fish Shellfish Immunol. 2018, 81, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Lu, Z.C.; Zhu, X.W.; Zhu, C.H.; Wang, C.G.; Shen, Y.C.; Wang, W. Differential expression of microRNAs in hemocytes from white shrimp Litopenaeus vannamei under copper stress. Fish Shellfish Immunol. 2018, 74, 152–161. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Kim, V.N. RNA interference in functional genomics and medicine. J. Korean Med. Sci. 2003, 18, 309–318. [Google Scholar] [CrossRef]
- Xu, J.; Han, F.; Zhang, X. Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA. Antiviral Res. 2007, 73, 126–131. [Google Scholar] [CrossRef]
- Story, B.; Ma, X.; Ishihara, K.; Li, H.; Hall, K.; Peak, A.; Anoja, P.; Park, J.; Haug, J.; Blanchette, M.; et al. Defining the expression of PiRNA and transposable elements in Drosophila ovarian germline stem cells and somatic support cells. Life Sci. Alliance 2019, 2, 1–16. [Google Scholar] [CrossRef]
- Pillai, R.S.; Chuma, S. PiRNAs and their involvement in male germline development in mice. Dev. Growth Differ. 2012, 54, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R. Dynamics of transposable element invasions with piRNA clusters. Mol. Biol. Evol. 2019, 36, 1457–1472. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Tóth, K.F. Piwi induces PiRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defense against transposable elements. Nat. Ecol. Evol. 2017, 2, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Waiho, K.; Fazhan, H.; Zhang, Y.; Li, S.; Zhang, Y.; Zheng, H.; Ikhwanuddin, M.; Ma, H. Comparative profiling of ovarian and testicular PiRNAs in the mud crab Scylla paramamosain. Genomics 2020, 112, 323–331. [Google Scholar] [CrossRef]
- Jehn, J.; Gebert, D.; Pipilescu, F.; Stern, S.; Kiefer, J.S.T.; Hewel, C.; Rosenkranz, D. PIWI genes and miRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun. Biol. 2018, 1, 137. [Google Scholar] [CrossRef]
- ter Horst, A.M.; Nigg, J.C.; Dekker, F.M.; Falk, B.W. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-interacting RNAs. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef]
- Huang, S.; Yoshitake, K.; Asakawa, S. A review of discovery profiling of piwi-interacting RNAs and their diverse functions in metazoans. Int. J. Mol. Sci. 2021, 22, 11166. [Google Scholar] [CrossRef]
- Taengchaiyaphum, S.; Wongkhaluang, P.; Sittikankaew, K.; Karoonuthaisiri, N.; Flegel, T.W.; Sritunyalucksana, K. Shrimp genome sequence contains independent clusters of ancient and current endogenous viral elements (EVE) of the parvovirus IHHNV. BMC Genom. 2022, 23, 565. [Google Scholar] [CrossRef]
- Huerlimann, R.; Wade, N.M.; Gordon, L.; Montenegro, J.D.; Goodall, J.; McWilliam, S.; Tinning, M.; Siemering, K.; Giardina, E.; Donovan, D.; et al. De novo assembly, characterization, functional annotation and expression patterns of the black tiger shrimp (Penaeus Monodon) transcriptome. Sci. Rep. 2018, 8, 13553. [Google Scholar] [CrossRef]
- Liu, D.; Hong, Z.; Gui, L.; Zhao, L.; Wang, Y.; Sun, S.; Li, M. Full-length transcriptomes and sex-based differentially expressed genes in the brain and ganglia of giant river prawn Macrobrachium rosenbergii. Biomolecules 2023, 13, 460. [Google Scholar] [CrossRef] [PubMed]
- Taneerat, C.; Olasard, P.; Suksri, P.; Whankaew, S.; Sathapondecha, P. Identification and profiling of long non-coding RNAs during molt cycle: An involvement of Lnc1182 in the molt of white shrimp, Litopenaeus vannamei. Aquac. Rep. 2023, 30, 101611. [Google Scholar] [CrossRef]
- Machado, M.; Fernández-Boo, S.; Teixeira, C.; Viegas, M.; Serradeiro, R.; Dias, J.; Costas, B.; Masagounder, K. DL-Methionyl-DL-Methionine as an efficient methionine source for promoting zootechnical performance and methionine-related pathways in the whiteleg shrimp (Penaeus vannamei). Br. J. Nutr. 2023, 130, 616–632. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, Y.; Zhang, Y.; Xu, B.; Sagada, G.; Wang, Z.; Chen, C.; Lang, X.; Zhang, J.; Shao, Q. Comparative study on the effects of crystalline L-methionine and methionine hydroxy analogue calcium supplementations in the diet of juvenile pacific white shrimp (Litopenaeus vannamei). Front. Physiol. 2023, 14, 105. [Google Scholar] [CrossRef]
- Asaikkutti, A.; Bhavan, P.S.; Vimala, K. Effects of different levels of dietary folic acid on the growth performance, muscle composition, immune response and antioxidant capacity of freshwater prawn, Macrobrachium rosenbergii. Aquaculture 2016, 464, 136–144. [Google Scholar] [CrossRef]
- Marçal, R.; Llorente, L.; Herrero, O.; Planelló, R.; Guilherme, S.; Pacheco, M. Intergenerational patterns of DNA methylation in Procambarus clarkii following exposure to genotoxicants: A conjugation in past simple or past continuous? Toxics 2021, 9, 271. [Google Scholar] [CrossRef]
- Mirbahai, L.; Chipman, J.K. Epigenetic memory of environmental organisms: A reflection of lifetime stressor exposures. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 764–765, 10–17. [Google Scholar] [CrossRef]
- Šrut, M. Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere 2021, 282, 131026. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, J.; Su, Y.; Liu, Z.; Mao, Y. Air exposure affects physiological responses, innate immunity, apoptosis and DNA methylation of kuruma shrimp, Marsupenaeus japonicus. Front. Physiol. 2020, 11, 223. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Lei, Y.; Zeng, Q.; Tan, G.; Yuan, Z.; Zhang, N.; Liu, J.; Wang, W. A first glimpse into the M6A modification machinery of shrimp: Genomic features, expression patterns and potential roles in molting regulation. Aquac. Rep. 2023, 29, 101493. [Google Scholar] [CrossRef]
- Yang, P.; Aweya, J.J.; Yao, D.; Wang, F.; Lun, J.; Hong, Y.; Sun, K.; Zhang, Y. The Krüppel-like factor of Penaeus vannamei negatively regulates transcription of the small subunit hemocyanin gene as part of shrimp immune response. Fish Shellfish Immunol. 2020, 100, 397–406. [Google Scholar] [CrossRef]
- Hou, F.; He, S.; Liu, Y.; Zhu, X.; Sun, C.; Liu, X. RNAi knock-down of shrimp Litopenaeus vannamei Toll gene and immune deficiency gene reveals their difference in regulating antimicrobial peptides transcription. Dev. Comp. Immunol. 2014, 44, 255–260. [Google Scholar] [CrossRef]
- Amparyup, P.; Sutthangkul, J.; Charoensapsri, W.; Tassanakajon, A. Pattern recognition protein binds to lipopolysaccharide and β-1,3-glucan and activates shrimp prophenoloxidase system. J. Biol. Chem. 2012, 287, 10060–10069. [Google Scholar] [CrossRef]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, J.; Xiang, J. A CRISPR/Cas9-mediated mutation in chitinase changes immune response to bacteria in Exopalaemon carinicauda. Fish Shellfish Immunol. 2017, 71, 43–49. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Yuan, J.; Zhang, C.; Li, S.; Li, F. CRISPR/Cas9-mediated mutation on an insulin-like peptide encoding gene affects the growth of the ridgetail white prawn Exopalaemon carinicauda. Front. Endocrinol. 2022, 13, 986491. [Google Scholar] [CrossRef]
- Gavery, M.R.; Roberts, S.B. Epigenetic considerations in aquaculture. PeerJ 2017, 5, e4147. [Google Scholar] [CrossRef]
- Norouzitallab, P.; Baruah, K.; Lulijwa, R.; Sorgeloos, P.; Bossier, P.; Vanrompay, D. Epigenetic Management of Disease Resistance in Farmed Shrimp; 5m books Ltd.: Essex, UK, 2022; ISBN 9781789181043. [Google Scholar]
- Xu, Y.; Liu, H.; Han, D.; Ren, L.; Gong, X.; Jiang, F.; Cui, Y.; Liu, X.; Ren, C.; Xue, J.; et al. Metabolomic alterations in the digestive system of the mantis shrimp Oratosquilla oratoria following short-term exposure to cadmium. Front. Physiol. 2021, 12, 706579. [Google Scholar] [CrossRef]
- Koch, I.J.; Nuetzel, H.M.; Narum, S.R. Epigenetic effects associated with salmonid supplementation and domestication. Environ. Biol. Fishes 2022, 106, 1093–1111. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, M.N.; Panserat, S.; Dupont-Nivet, M.; Quillet, E.; Montfort, J.; Le Cam, A.; Medale, F.; Kaushik, S.J.; Geurden, I. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genom. 2016, 17, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Panserat, S.; Marandel, L.; Geurden, I.; Veron, V.; Dias, K.; Plagnes-Juan, E.; Pegourié, G.; Arbenoits, E.; Santigosa, E.; Weber, G.; et al. Muscle catabolic capacities and global hepatic epigenome are modified in juvenile rainbow trout fed different vitamin levels at first feeding. Aquaculture 2017, 468, 515–523. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, Q.; Yu, H.; Kong, L.F. Genetic and epigenetic variation in mass selection populations of Pacific oyster Crassostrea gigas. Genes Genom. 2013, 35, 641–647. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wikumpriya, G.C.; Prabhatha, M.W.S.; Lee, J.; Kim, C.-H. Epigenetic Modulations for Prevention of Infectious Diseases in Shrimp Aquaculture. Genes 2023, 14, 1682. https://doi.org/10.3390/genes14091682
Wikumpriya GC, Prabhatha MWS, Lee J, Kim C-H. Epigenetic Modulations for Prevention of Infectious Diseases in Shrimp Aquaculture. Genes. 2023; 14(9):1682. https://doi.org/10.3390/genes14091682
Chicago/Turabian StyleWikumpriya, Gunasekara Chathura, Madhuranga Walawedurage Srinith Prabhatha, Jiye Lee, and Chan-Hee Kim. 2023. "Epigenetic Modulations for Prevention of Infectious Diseases in Shrimp Aquaculture" Genes 14, no. 9: 1682. https://doi.org/10.3390/genes14091682
APA StyleWikumpriya, G. C., Prabhatha, M. W. S., Lee, J., & Kim, C. -H. (2023). Epigenetic Modulations for Prevention of Infectious Diseases in Shrimp Aquaculture. Genes, 14(9), 1682. https://doi.org/10.3390/genes14091682