Dual RNA-Seq Analysis of Trichophyton rubrum and HaCat Keratinocyte Co-Culture Highlights Important Genes for Fungal-Host Interaction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
2.2. Keratinocytes, Media and Growth Conditions
2.3. Co-Culture Assay and Conditions
2.4. RNA Isolation and Integrity Analysis
2.5. Library Construction and Sequencing
2.6. Sequence Data Analysis
2.7. qPCR Validation
3. Results
3.1. Electron Microscopy of T. rubrum and HaCat Co-Culture
3.2. Dual RNA-Seq Analysis of the Fungal-Host Interaction
3.3. Transcriptional Profile Analysis of Differentially Expressed Genes in the T. rubrum-Keratinocyte Co-Culture System
3.4. Functional Categorization of Differentially Expressed Genes
3.5. Validation by qPCR
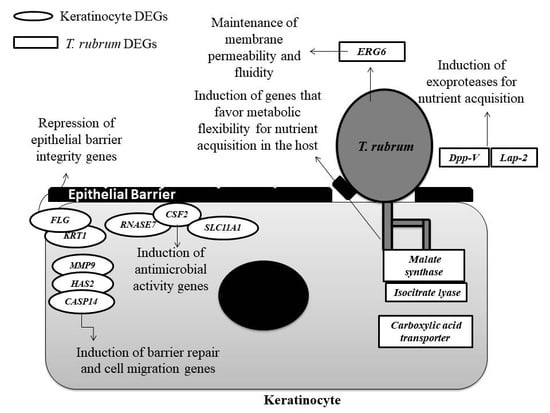
4. Discussion
4.1. Genes Involved in Protease Secretion Are Important for the Pathogenicity of T. rubrum
4.2. The ERG6 Gene Is a Promising Target for Developing a New Antifungal Agent Against T. rubrum
4.3. Glyoxylate Cycle Genes and a Carboxylic Acid Transporter May Be Associated with Mechanisms of Metabolic Flexibility in the T. rubrum-Host Relationship
4.4. The Modulation of Genes Involved in the Maintenance of the Skin Barrier, Cell Migration, and Differentiation May Be Associated with the Defense Strategies of Human Keratinocytes
4.5. The Induction of Genes Involved in the Immune Response of Human Keratinocytes that Encode Compounds with Antimicrobial Activity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bouchara, J.P.; Mignon, B.; Chaturvedi, V. Dermatophytes and dermatophytoses: a thematic overview of state of the art, and the directions for future research and developments. Mycopathologia 2017, 182, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Rodwell, G.E.; Bayles, C.L.; Towersey, L.; Aly, R. The prevalence of dermatophyte infection in patients infected with human immunodeficiency virus. Int. J. Dermatol. 2008, 47, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Romano, C.; Massai, L.; Asta, F.; Signorini, A.M. Prevalence of dermatophytic skin and nail infections in diabetic patients. Mycoses 2001, 44, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Aly, R. Ecology and epidemiology of dermatophyte infections. J. Am. Acad. Dermatol. 1994, 31, S21–S25. [Google Scholar] [CrossRef]
- Havlickova, B.; Czaika, V.A.; Fredrich, M. Epidemiological trends in skin mycosis worldwide. Mycosis 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Hube, B.; Hay, R.; Brasch, J.; Veraldi, S.; Schaller, M. Dermatomycoses and inflammation: The adaptive balance between growth, damage, and survival. J. Mycol. Med. 2015, 25, e44–e58. [Google Scholar] [CrossRef] [PubMed]
- Achterman, R.R.; White, T.C. A foot in the door for dermatophyte research. PLoS Pathog. 2012, 8, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Kämmer, P.; Brunke, S.; Linde, J. Two’s company: Studying interspecies relationships with dual RNA-seq. Curr. Opin. Microbiol. 2018, 42, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Aprianto, R.; Slager, J.; Holsappel, S.; Veening, J.-W. Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol. 2016, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Wesolowska-Andersen, A.; Everman, J.L.; Davidson, R.; Rios, C.; Herrin, R.; Eng, C.; Janssen, W.J.; Liu, A.H.; Oh, S.S.; Kumar, R.; et al. Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome. Genome Biol. 2017, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Tierney, L.; Linde, J.; Müller, S.; Brunke, S.; Molina, J.C.; Hube, B.; Schöck, U.; Guthke, R.; Kuchler, K. An interspecies regulatory network inferred from simultaneous RNA-seq of Candida albicans invading innate immune cells. Front. Microbiol. 2012, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.E.; Shuey, L.S.; Naidoo, S.; Mamni, T.; Berger, D.K.; Myburg, A.A.; Van den Berg, N.; Naidoo, S. Dual RNA-sequencing of Eucalyptus nitens during Phytophthora cinnamomi challenge reveals pathogen and host factors influencing compatibility. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Komoto, T.T.; Bitencourt, T.A.; Silva, G.; Beleboni, R.O.; Marins, M.; Fachin, A.L. Gene expression response of Trichophyton rubrum during coculture on keratinocytes exposed to antifungal agents. Evid. Based Complement. Altern. Med. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Santiago, K.; Bomfim, G.F.; Criado, P.R.; Almeida, S.R. Monocyte-derived dendritic cells from patients with dermatophytosis restrict the growth of Trichophyton rubrum and induce CD4-T cell activation. PLoS ONE 2014, 9, e110879. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.A.; Harris, M.A. The Gene Ontology (GO) Project: Structured vocabularies for molecular biology and their application to genome and expression analysis. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 1–9. ISBN 0471250953. [Google Scholar]
- Gotz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vêncio, R.Z.N.; Koide, T.; Gomes, S.L.; de Pereira, C.A.B. BayGO: Bayesian analysis of ontology term enrichment in microarray data. BMC Bioinf. 2006, 7, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zhang, D.; Hu, P.-C.; Li, Q.; Lin, C.-Y. HOXB7-S3 inhibits the proliferation and invasion of MCF-7 human breast cancer cells. Mol. Med. Rep. 2015, 4901–4908. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Ma, X.; Kang, H.; Gao, J.; Min, W.; Guan, H.; Diao, Y.; Lu, W.; Wang, X. Antitumor activity of the selective cyclooxygenase-2 inhibitor, celecoxib, on breast cancer in vitro and in vivo. Cancer Cell Int. 2012, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Komoto, T.T.; Massaroto, B.G.; Miranda, C.E.S.; Beleboni, R.O.; Marins, M.; Fachin, A.L. Trans-chalcone and quercetin down-regulate fatty acid synthase gene expression and reduce ergosterol content in the human pathogenic dermatophyte Trichophyton rubrum. BMC Complement. Altern. Med. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Jacob, T.R.; Peres, N.T.A.; Persinoti, G.F.; Silva, L.G.; Mazucato, M.; Rossi, A.; Martinez-Rossi, N.M. Rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 2012, 50, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Barquist, L.; Vogel, J. Resolving host–pathogen interactions by dual RNA-seq. PLoS Pathog. 2017, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Oono, Y.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Minami, E. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 2012, 7, e49423. [Google Scholar] [CrossRef] [PubMed]
- Abadio, A.K.R.; Kioshima, E.S.; Teixeira, M.M.; Martins, N.F.; Maigret, B.; Felipe, M.S.S. Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC Genom. 2011, 12, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldo, A.; Monod, M.; Mathy, A.; Cambier, L.; Bagut, E.T.; Defaweux, V.; Symoens, F.; Antoine, N.; Mignon, B. Mechanisms of skin adherence and invasion by dermatophytes. Mycoses 2012, 55, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Peres, N.T.D.A.; Maranhão, F.C.A.; Rossi, A.; Martinez-Rossi, N.M. Dermatophytes: Host-pathogen interaction and antifungal resistance. An. Bras. Dermatol. 2010, 85, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Macedo, C.; Franco, M.E.; Assis, A.F.; Komoto, T.T.; Stehling, E.G.; Beleboni, R.O.; Malavazi, I.; Marins, M.; Fachin, A.L. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genom. 2016, 17, 249. [Google Scholar] [CrossRef] [PubMed]
- Monod, M.; Léchenne, B.; Jousson, O.; Grand, D.; Zaugg, C.; Stöcklin, R.; Grouzmann, E. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 2005, 151, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Leng, W.; Liu, T.; Wang, J.; Li, R.; Jin, Q. Expression dynamics of secreted protease genes in Trichophyton rubrum induced by key host’s proteinaceous components. Med. Mycol. 2009, 47, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.S.; Abro, A.; Raza, S.; Saroosh, A. Structure and dynamics studies of sterol 24-C-methyltransferase with mechanism based inactivators for the disruption of ergosterol biosynthesis. Mol. Biol. Rep. 2014, 41, 4279–4293. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, T.; Iefuji, H.; Hiraga, Y.; Hosomi, A.; Morita, T.; Giga-Hama, Y.; Takegawa, K. Multiple functions of ergosterol in the fission yeast Schizosaccharomyces pombe. Microbiology 2008, 154, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.S.; Leitner, I.; Favre, B.; Neil, S.; Osborne, C.S.; Leitner, I.; Favre, B.; Ryder, N.S. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. 2005, 49, 2840–2844. [Google Scholar] [CrossRef] [PubMed]
- Gaber, R.F.; Copple, D.M.; Kennedy, B.K.; Vidal, M.; Bard, M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 1989, 9, 3447–3456. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheah, H.L.; Lim, V.; Sandai, D. Inhibitors of the glyoxylate cycle enzyme ICL1 in Candida albicans for potential use as antifungal agents. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Fleck, C.B.; Schöbel, F.; Brock, M. Nutrient acquisition by pathogenic fungi: Nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 2011, 301, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Cantelli, B.A.M.; Bitencourt, T.A.; Komoto, T.T.; Beleboni, R.O.; Marins, M.; Fachin, A.L. Caffeic acid and licochalcone A interfere with the glyoxylate cycle of Trichophyton rubrum. Biomed. Pharmacother. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.; Casal, M.; Johansson, B.; MacCallum, D.M.; Brown, A.J.P.; Paiva, S. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol. Microbiol. 2010, 75, 1337–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.J.; Kim, J.Y.; Song, C.H.; Jung, H.D.; Lee, S.H.; Lee, S.J.; Kim, D.W. Disruption of barrier function in dermatophytosis and pityriasis versicolor. J. Dermatol. 2011, 38, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Irvine, A.D. Atopic eczema and the filaggrin story. Semin. Cutan. Med. Surg. 2008, 27, 128–137. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.A. Filaggrin and the great epidermal barrier grief. Australas. J. Dermatol. 2008, 49, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.-M.; Pfeiffer, S.; Akaki, T.; Schröder, J.-M.; Kleine, M.; Neumann, C.; Proksch, E.; Brasch, J. Barrier function, epidermal differentiation, and human β-defensin 2 expression in Tinea Corporis. J. Investig. Dermatol. 2007, 127, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Roth, W.; Kumar, V.; Beer, H.-D.; Richter, M.; Wohlenberg, C.; Reuter, U.; Thiering, S.; Staratschek-Jox, A.; Hofmann, A.; Kreusch, F.; et al. Keratin 1 maintains skin integrity and participates in an inflammatory network in skin through interleukin-18. J. Cell Sci. 2012, 125, 5269–5279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, W.; Wilson, C.; López-Boado, Y. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Purwar, R.; Kraus, M.; Werfel, T.; Wittmann, M. Modulation of keratinocyte-derived MMP-9 by IL-13: A possible role for the pathogenesis of epidermal inflammation. J. Investig. Dermatol. 2008, 128, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hvid, M.; Johansen, C.; Deleuran, B.; Kemp, K.; Deleuran, M.; Vestergaard, C. Regulation of caspase 14 expression in keratinocytes by inflammatory cytokines—A possible link between reduced skin barrier function and inflammation? Exp. Dermatol. 2011, 20, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Gkegkes, I.D.; Aroni, K.; Agrogiannis, G.; Patsouris, E.S.; Konstantinidou, A.E. Expression of caspase-14 and keratin-19 in the human epidermis and appendages during fetal skin development. Arch. Dermatol. Res. 2013, 305, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Lippens, S.; Kockx, M.; Knaapen, M.; Mortier, L.; Polakowska, R.; Verheyen, A.; Garmyn, M.; Zwijsen, A.; Formstecher, P.; Huylebroeck, D.; et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 2000, 7, 1218–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denecker, G.; Hoste, E.; Gilbert, B.; Hochepied, T.; Ovaere, P.; Lippens, S.; Van den Broecke, C.; Van Damme, P.; D’Herde, K.; Hachem, J.-P.; et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell Biol. 2007, 9, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Ratzinger, G.; Stoitzner, P.; Ebner, S.; Lutz, M.B.; Layton, G.T.; Rainer, C.; Senior, R.M.; Shipley, J.M.; Fritsch, P.; Schuler, G.; et al. Matrix metalloproteinases 9 and 2 are necessary for the migration of langerhans cells and dermal dendritic cells from human and murine skin. J. Immunol. 2002, 168, 4361–4371. [Google Scholar] [CrossRef] [PubMed]
- Visse, R.; Nagase, H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: Structure, function, and biochemistry. Circ. Res. 2003, 92, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zheng, M.; Kim, B.; Rouse, B.T. Role of matrix metalloproteinase-9 in angiogenesis caused by ocular infection with herpes simplex virus. J. Clin. Investig. 2002, 110, 1105–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, T.M.; Passos, S.T.; Novais, F.O.; Beiting, D.P.; Costa, R.S.; Queiroz, A.; Mosser, D.; Scott, P.; Carvalho, E.M.; Carvalho, L.P. Matrix metalloproteinase 9 production by monocytes is enhanced by TNF and participates in the pathology of human Cutaneous Leishmaniasis. PLoS Negl. Trop. Dis. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Aspects Med. 2009, 29, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Liang, J.; Noble, P.W. Hyaluronan in tissue injury and repair. Annu. Rev. Cell Dev. Biol. 2007, 23, 435–461. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Akifusa, S.; Itano, N.; Kimata, K.; Kawamura, T.; Koseki, T.; Takehara, T.; Nishihara, T. Potential role of high molecular weight hyaluronan in the anti-Candida activity of human oral epithelial cells. Med. Mycol. 2007, 45, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Becknell, B.; Spencer, J.D. A Review of ribonuclease 7’s structure, regulation, and contributions to host defense. Int. J. Mol. Sci. 2016, 17, 423. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [PubMed]
- Fritz, P.; Beck-Jendroschek, V.; Brasch, J. Inhibition of dermatophytes by the antimicrobial peptides human β-defensin-2, ribonuclease 7 and psoriasin. Med. Mycol. 2012, 50, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehra, T.; Köberle, M.; Braunsdorf, C.; Mailänder-Sanchez, D.; Borelli, C.; Schaller, M. Alternative approaches to antifungal therapies. Exp. Dermatol. 2012, 21, 778–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubel, K.; Dale, D.C.; Liles, W.C. Therapeutic use of cytokines to modulate phagocyte function for the treatment of infectious diseases: Current status of granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, macrophage colony-stimulating factor, and interferon-gamma. J. Infect. Dis. 2002, 185, 1490–1501. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, C.H.; Roberts, A.I.; Das, J.; Xu, G.; Ren, G.; Zhang, Y.; Zhang, L.; Yuan, Z.R.; Tan, H.S.W.; et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: What we do and don’t know. Cell Res. 2006, 16, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Liehl, E.; Hildebrandt, J.; Lam, C.; Mayer, P. Prediction of the role of granulocyte-macrophage colony-stimulating factor in animals and man from in vitro results. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13, 9–17. [Google Scholar] [CrossRef]
- Bodey, G.P.; Anaissie, E.; Gutterman, J.; Vadhan-Raj, S. Role of granulocyte-macrophage colony-stimulating factor as adjuvant treatment in neutropenic patients with bacterial and fungal infection. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13 (Suppl. 2), S18–S22. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.A. GM-CSF in inflammation and autoimmunity. Trends Immunol. 2002, 23, 403–408. [Google Scholar] [CrossRef]
- Rademacher, F.; Simanski, M.; Harder, J. RNase 7 in cutaneous defense. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Nairz, M.; Fritsche, G.; Crouch, M.L.V.; Barton, H.C.; Fang, F.C.; Weiss, G. Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell. Microbiol. 2009, 11, 1365–1381. [Google Scholar] [CrossRef] [PubMed]
- Stober, C.B.; Brode, S.; White, J.K.; Popoff, J.-F.; Blackwell, J.M. Slc11a1, formerly Nramp1, is expressed in dendritic cells and influences major histocompatibility complex class II expression and antigen-presenting cell function. Infect. Immun. 2007, 75, 5059–5067. [Google Scholar] [CrossRef] [PubMed]
- Noronha, S.A.; Noronha, S.M.; Lanziani, L.E.; Ferreira, L.M.; Gragnani, A. Innate and adaptive immunity gene expression of human keratinocytes cultured of severe burn injury. Acta Cir. Bras. 2014, 29 (Suppl. 3), 60–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackwell, J.M.; Searle, S.; Goswami, T.; Miller, E.N. Understanding the multiple functions of Nrampl. Microbes Infect. 2000, 2, 317–321. [Google Scholar] [CrossRef]
- Blackwell, J.M.; Goswami, T.; Evans, C.A.W.; Sibthorpe, D.; Papo, N.; White, J.K.; Searle, S.; Miller, E.N.; Peacock, C.S.; Mohammed, H.; et al. SLC11A1 (formerly NRAMP1) and disease resistance. Cell. Microbiol. 2001, 3, 773–784. [Google Scholar] [CrossRef] [PubMed]




| ID | Gene Product Name | Log2 Fold Change |
|---|---|---|
| SLC9A2 | Sodium/hydrogen exchanger 2 | 5.01 |
| ANGPTL4 | Angiopoietin-related protein 4 | 4.71 |
| DES | Desmin | 4.53 |
| C4orf47 | UPF0602 protein C4orf47 | 4.51 |
| KISS1R | KiSS-1 receptor | 4.49 |
| NSA2 | Ribosome biogenesis protein NSA2 homolog | 4.35 |
| HIST1H3C | Histone cluster 1 H3 family member c | 4.04 |
| SEC11C | Signal peptidase complex catalytic subunit | 3.87 |
| KPNA7 | Importin subunit alpha-8 | 3.83 |
| CASP14 | Caspase 14 | 3.74 |
| SLC2A3 | Facilitated glucose transporter member 3 | 3.73 |
| ALDOC | Fructose-bisphosphate aldolase C | 3.70 |
| MT1B | Metallothionein-1B | 3.62 |
| SERPINE1 | Plasminogen activator inhibitor 1 | 3.55 |
| MAF | Transcription factor Maf | 3.54 |
| CA9 | Carbonic anhydrase 9 | 3.36 |
| TGM2 | Transglutaminase 2 | 3.35 |
| PADI1 | Protein-arginine deiminase type-1 | 3.29 |
| STC1 | Stanniocalcin 1 | 3.14 |
| BNIP3 | BCL2 interacting protein 3 | 3.08 |
| LSS | Lanosterol synthase | 3.06 |
| MT1H | Metallothionein 1H | 3.05 |
| MT1X | Metallothionein 1X | 2.97 |
| PLA2G2F | Group IIF secretory phospholipase A2 | 2.96 |
| CALB1 | Calbindin 1 | 2.93 |
| POTEM | Putative POTE ankyrin domain family member M | −5.31 |
| SNORA51 | Small nucleolar RNA. H/ACA box | −4.90 |
| ANP32A-IT1 | ANP32A intronic transcript 1 | −4.64 |
| UCKL1 | Uridine-cytidine kinase 1 like 1 | −4.50 |
| FNDC3B | Fibronectin type III domain containing | −4.37 |
| KRT1 | Keratin 1 | −4.02 |
| MMP12 | Matrix metallopeptidase 12 | −3.22 |
| NSD1 | Nuclear receptor binding SET domain | −3.06 |
| CYCSP52 | Cytochrome c. somatic pseudogene | −3.02 |
| EME2 | Essential meiotic structure-specific endonuclease subunit 2 | −3.00 |
| COL12A1 | Collagen type XII alpha 1 chain | −2.88 |
| SNORD45A | Small nucleolar RNA. C/D box | −2.82 |
| FBXL19-AS1 | FBXL19 antisense RNA 1 (head to head) | −2.80 |
| TRIM26 | Tripartite motif containing 26 | −2.76 |
| IARS | Isoleucyl-tRNA synthetase | −2.76 |
| KIF14 | Kinesin family member 14 | −2.74 |
| MEGF8 | Multiple EGF like domains 8 | −2.67 |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein | −2.66 |
| ID | Gene Product Name | Log2 Fold Change |
|---|---|---|
| TERG_12606 | Dipeptidyl peptidase V (DPPV) | 2.16 |
| TERG_01280 | Hypothetical protein | 2.06 |
| TERG_03102 | Sterol 24-C-methyltransferase- ERG6 | 2.05 |
| TERG_08104 | Potassium/sodium efflux P-type ATPase | 1.98 |
| TERG_01281 | Malate synthase | 1.72 |
| TERG_04399 | Phthalate transporter | 1.62 |
| TERG_00215 | MFS peptide transporter | 1.47 |
| TERG_00348 | Galactose-proton symporter | 1.47 |
| TERG_02811 | Hypothetical protein | 1.42 |
| TERG_12645 | Hypothetical protein | 1.40 |
| TERG_07017 | Oxidoreductase | 1.35 |
| TERG_08333 | 1-pyrroline-5-carboxylate dehydrogenase | 1.34 |
| TERG_02671 | Hypothetical protein | 1.34 |
| TERG_02023 | Extracellular matrix protein | 1.32 |
| TERG_08405 | Leucine aminopeptidase 2 | 1.30 |
| TERG_00916 | Carboxylic acid transporter | 1.29 |
| TERG_11638 | Isocitrate lyase | 1.28 |
| TERG_04952 | ABC transporter | 1.26 |
| TERG_01406 | Hypothetical protein | −2.91 |
| TERG_07726 | Hypothetical protein | −2.25 |
| TERG_03174 | MFS siderochrome iron transporter | −1.99 |
| TERG_06355 | Hypothetical protein | −1.90 |
| TERG_07035 | Hypothetical protein | −1.85 |
| TERG_04156 | Hypothetical protein | −1.77 |
| TERG_05655 | AN1 zinc finger protein | −1.73 |
| TERG_01622 | Hypothetical protein | −1.63 |
| TERG_07477 | Hypothetical protein | −1.57 |
| TERG_06186 | Protein disulfide-isomerase domain-containing protein | −1.57 |
| TERG_03708 | Hypothetical protein | −1.53 |
| TERG_03855 | Hypothetical protein | −1.50 |
| TERG_00499 | Hypothetical protein | −1.45 |
| TERG_03175 | Hypothetical protein | −1.45 |
| TERG_04073 | Glutathione synthetase | −1.41 |
| TERG_12563 | Hypothetical protein | −1.37 |
| TERG_08139 | NAD dependent epimerase/dehydratase | −1.34 |
| TERG_06963 | Hsp90-like protein | −1.33 |
| TERG_01731 | Hypothetical protein | −1.32 |
| TERG_04006 | Rho guanyl nucleotide exchange factor | −1.32 |
| ID | Gene Product Name | Log2 Fold Change |
|---|---|---|
| Metabolic process | ||
| TERG_03102 | Sterol 24-C-methyltransferase | 2.05 |
| TERG_08104 | Sodium transport ATPase | 1.98 |
| TERG_02811 | Hypothetical protein | 1.40 |
| TERG_08333 | Delta 1-pyrroline-5-carboxylate dehydrogenase | 1.34 |
| TERG_11638 | Isocitrate lyase | 1.26 |
| TERG_01270 | AMP-dependent ligase | 1.13 |
| TERG_07691 | Nonspecific lipid-transfer protein | 1.13 |
| TERG_07222 | Carbonic anhydrase | 1.05 |
| Transmembrane transport | ||
| TERG_04399 | Phthalate transporter (MFS transporter) | 1.62 |
| TERG_00348 | Galactose-proton symporter (MFS transporter) | 1.42 |
| TERG_00916 | Carboxylic acid transporter (MFS transporter) | 1.28 |
| TERG_04952 | ABC transporter | 1.25 |
| TERG_04356 | Amino acid permease | 1.06 |
| Pathogenesis | ||
| TERG_12606 | Dipeptidyl peptidase V | 2.16 |
| TERG_08405 | Leucine Aminopeptidase 2 | 1.29 |
| Glyoxylate cycle | ||
| TERG_01281 | Malate synthase | 1.72 |
| TERG_11638 | Isocitrate lyase | 1.26 |
| TERG_11639 | Isocitrate lyase | 1.13 |
| ID | Gene Product Name | Log2 Fold Change |
|---|---|---|
| Positive regulation of cell migration | ||
| TCAF2 | TRPM8 channel-associated factor 2 | 2.11 |
| MMP9 | Matrix metalloproteinase-9 | 2.06 |
| LAMC2 | Laminin subunit gamma-2 | 1.97 |
| HBEGF | Proheparin-binding EGF-like growth factor | 1.84 |
| HAS2 | Hyaluronan synthase 2 | 1.46 |
| MAPK cascade involved in the innate immune response | ||
| CSF2 | Granulocyte-macrophage colony-stimulating factor | 2.86 |
| HBEGF | Proheparin-binding EGF-like growth factor | 1.84 |
| DUSP5 | Dual specificity protein phosphatase 5 | 1.57 |
| PSMB3 | Proteasome subunit beta type-3 | 1.46 |
| PPP5C | Serine/threonine-protein phosphatase 5 | 1.37 |
| PSMB2 | Proteasome subunit beta type-2 | 1.28 |
| UBB | Polyubiquitin-B | 1.24 |
| Antimicrobial humoral immune response | ||
| SERPINE1 | Plasminogen activator inhibitor 1 | 3.55 |
| SLC11A1 | Natural resistance-associated macrophage protein 1 | 2.28 |
| RNASE7 | Ribonuclease 7 | 2.27 |
| RPS19 | 40S ribosomal protein S19 | 1.66 |
| RPL30 | 60S ribosomal protein L30 | 1.41 |
| Epidermal cell differentiation | ||
| CASP14 | Caspase-14 | 3.74 |
| ALDOC | Fructose-bisphosphate aldolase C | 3.70 |
| AKR1C1 | Aldo-keto reductase family | 2.80 |
| LAMC2 | Laminin subunit gamma-2 | 1.97 |
| PGK1 | Phosphoglycerate kinase 1 | 1.62 |
| Establishment of the skin barrier | ||
| KRT1 | Keratin type II cytoskeletal 1 | −4.02 |
| FLG | Filaggrin | −1.86 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrucelli, M.F.; Peronni, K.; Sanches, P.R.; Komoto, T.T.; Matsuda, J.B.; Silva, W.A.d., Jr.; Beleboni, R.O.; Martinez-Rossi, N.M.; Marins, M.; Fachin, A.L. Dual RNA-Seq Analysis of Trichophyton rubrum and HaCat Keratinocyte Co-Culture Highlights Important Genes for Fungal-Host Interaction. Genes 2018, 9, 362. https://doi.org/10.3390/genes9070362
Petrucelli MF, Peronni K, Sanches PR, Komoto TT, Matsuda JB, Silva WAd Jr., Beleboni RO, Martinez-Rossi NM, Marins M, Fachin AL. Dual RNA-Seq Analysis of Trichophyton rubrum and HaCat Keratinocyte Co-Culture Highlights Important Genes for Fungal-Host Interaction. Genes. 2018; 9(7):362. https://doi.org/10.3390/genes9070362
Chicago/Turabian StylePetrucelli, Monise Fazolin, Kamila Peronni, Pablo Rodrigo Sanches, Tatiana Takahasi Komoto, Josie Budag Matsuda, Wilson Araújo da Silva, Jr., Rene Oliveira Beleboni, Nilce Maria Martinez-Rossi, Mozart Marins, and Ana Lúcia Fachin. 2018. "Dual RNA-Seq Analysis of Trichophyton rubrum and HaCat Keratinocyte Co-Culture Highlights Important Genes for Fungal-Host Interaction" Genes 9, no. 7: 362. https://doi.org/10.3390/genes9070362
APA StylePetrucelli, M. F., Peronni, K., Sanches, P. R., Komoto, T. T., Matsuda, J. B., Silva, W. A. d., Jr., Beleboni, R. O., Martinez-Rossi, N. M., Marins, M., & Fachin, A. L. (2018). Dual RNA-Seq Analysis of Trichophyton rubrum and HaCat Keratinocyte Co-Culture Highlights Important Genes for Fungal-Host Interaction. Genes, 9(7), 362. https://doi.org/10.3390/genes9070362