Review of the Distribution and Influence of Antibiotic Resistance Genes in Ballast Water
Abstract
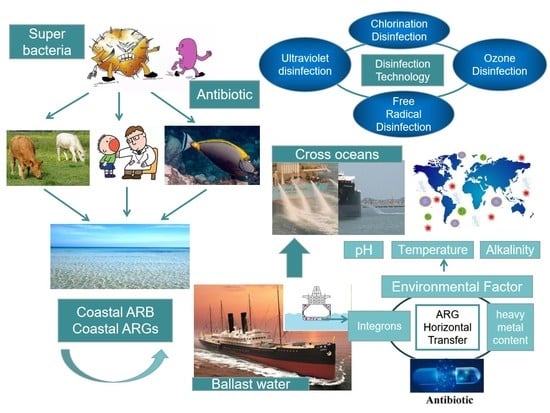
:1. Introduction
2. Methodology
2.1. Methods
2.2. Identifification of Records
2.3. Eligibility Criteria
3. Bacteria in Ship Ballast Water
4. Resistance Genes
4.1. Antibiotics and Antibiotic Resistance Genes
4.2. ARGs in the Marine Environment
5. ARGs in the Ballast Water
5.1. ARGs in the Ballast Water of Ships
5.2. ARGs in Ballast Water Sediments
5.3. Influencing Factors of Resistance Genes
6. Changes in ARGs in Ballast Water after Disinfection
6.1. Ballast Water Disinfection
6.2. Changes in ARGs in Ballast Water under Different Disinfection Technology
6.2.1. Chlorination Disinfection
6.2.2. Ultraviolet Disinfection
6.2.3. Ozone Disinfection
6.2.4. Free Radical Disinfection
6.3. The Affect of Subinhibitory Concentrations of Disinfectants
7. Conclusions
- Several resistance genes have been discovered in ballast water and sediments. This is generally due to the widespread use of antibiotics in aquaculture;
- The levels of ARGs in ballast water were higher than those in the samples collected from nearby marine environments. ARGs were found in ballast water samples from different sea areas. This result indicated that ballast water could promote the spread of ARGs, which should be further considered in the formulation of ballast water discharge standards;
- Disinfection treatment can enhance the removal of ARGs in ballast water. However, if the disinfectants are reduced to subinhibitory levels, a potential mechanism for the conjugate transfer of the ARGs will emerge.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lv, B.; Shi, J.; Li, T.; Ren, L.; Tian, W.; Lu, X.; Han, Y.; Cui, Y.; Jiang, T. Deciphering the characterization, ecological function and assembly processes of bacterial communities in ship ballast water and sediments. Sci. Total Environ. 2022, 816, 152721. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Q.; Xue, J.Z.; Xiao, N.Y.; Lv, B.Y.; Wu, H.X. Effects of holding time on the diversity and composition of potential pathogenic bacteria in ship ballast water. Mar. Environ. Res. 2020, 160, 104979. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Saebi, M.; Grey, E.K.; Corbett, J.J.; Chen, D.; Yang, D.; Wan, Z. Ballast water-mediated species spread risk dynamics and policy implications to reduce the invasion risk to the Mediterranean Sea. Mar. Pollut. Bull. 2022, 174, 113285. [Google Scholar] [CrossRef] [PubMed]
- Darling, J.A.; Martinson, J.; Gong, Y.G.; Oskum, S.; Pilgrim, E.; Lohan, K.M.P.; Carney, K.J.; Ruiz, G.M. Ballast Water Exchange and Invasion Risk Posed by Intracoastal Vessel Traffic: An Evaluation Using High Throughput Sequencing. Environ. Sci. Technol. 2018, 52, 9926–9936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gollasch, S.; David, M. Abiotic and biological differences in ballast water uptake and discharge samples. Mar. Pollut. Bull. 2021, 164, 112046. [Google Scholar] [CrossRef] [PubMed]
- Altug, G.; Gurun, S.; Cardak, M.; Ciftci, P.S.; Kalkan, S. The occurrence of pathogenic bacteria in some ships’ ballast water incoming from various marine regions to the Sea of Marmara, Turkey. Mar. Environ. Res. 2012, 81, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Werschkun, B.; Banerji, S.; Basurko, O.C.; David, M.; Fuhr, F.; Gollasch, S.; Grummt, T.; Haarich, M.; Jha, A.N.; Kacan, S.; et al. Emerging risks from ballast water treatment: The run-up to the International Ballast Water Management Convention. Chemosphere 2014, 112, 256–266. [Google Scholar] [CrossRef] [Green Version]
- Dong, P.Y.; Cui, Q.J.; Fang, T.T.; Huang, Y.; Wang, H. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J. Environ. Sci. 2019, 77, 65–74. [Google Scholar] [CrossRef]
- Cui, Q.J.; Huang, Y.; Wang, H.; Fang, T.T. Diversity and abundance of bacterial pathogens in urban rivers impacted by domestic sewage. Environ. Pollut. 2019, 249, 24–35. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Hartmann, A.; Alder, A.C.; Koller, T.; Widmer, R.M. Identification of fluoroquinolone antibiotics as the main source of umuC genotoxicity in native hospital wastewater. Environ. Toxicol. Chem. 1998, 17, 377–382. [Google Scholar] [CrossRef]
- Jain, R.; Rivera, M.C.; Moore, J.E.; Lake, J.A. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 2003, 20, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Schluter, A.; Szczepanowski, R.; Puhler, A.; Top, E.M. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 2007, 31, 449–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.K.; Ekka, R.; Mishra, M.; Mohapatra, H. Association study of multiple antibiotic resistance and virulence: A strategy to assess the extent of risk posed by bacterial population in aquatic environment. Environ. Monit. Assess. 2017, 189, 320. [Google Scholar] [CrossRef]
- Han, Y.; Wang, J.; Zhao, Z.L.; Chen, J.W.; Lu, H.; Liu, G.F. Fishmeal Application Induces Antibiotic Resistance Gene Propagation in Mariculture Sediment. Environ. Sci. Technol. 2017, 51, 10850–10860. [Google Scholar] [CrossRef]
- Ng, C.; Le, T.H.; Goh, S.G.; Liang, L.; Kim, Y.; Rose, J.B.; Yew-Hoong, K.G. A Comparison of Microbial Water Quality and Diversity for Ballast and Tropical Harbor Waters. PLoS ONE 2015, 10, e0154652. [Google Scholar] [CrossRef]
- Hoa, P.T.P.; Nonaka, L.; Viet, P.H.; Suzuki, S. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 2008, 405, 377–384. [Google Scholar] [CrossRef]
- Ju, F.; Li, B.; Ma, L.; Wang, Y.; Huang, D.; Zhang, T. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 2016, 91, 1–10. [Google Scholar] [CrossRef]
- Beeton, M.L.; Chalker, V.J.; Jones, L.C.; Maxwell, N.C.; Spiller, O.B. Antibiotic Resistance among Clinical Ureaplasma Isolates Recovered from Neonates in England and Wales between 2007 and 2013. Antimicrob. Agents Chemother. 2016, 60, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Song, W.; Lin, H.; Wang, W.; Du, L.; Xing, W. Antibiotics and antibiotic resistance genes in global lakes: A review and meta-analysis. Environ. Int. 2018, 116, 60–73. [Google Scholar] [CrossRef]
- Hatosy, S.M.; Martiny, A.C. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—Occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Parnanen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef] [Green Version]
- Cordero, O.X.; Wildschutte, H.; Kirkup, B.; Proehl, S.; Ngo, L.; Hussain, F.; Le Roux, F.; Mincer, T.; Polz, M.F. Ecological Populations of Bacteria Act as Socially Cohesive Units of Antibiotic Production and Resistance. Science 2012, 337, 1228–1231. [Google Scholar] [CrossRef]
- Lv, B.; Cui, Y.; Tian, W.; Li, J.; Xie, B.; Yin, F. Abundances and profiles of antibiotic resistance genes as well as co-occurrences with human bacterial pathogens in ship ballast tank sediments from a shipyard in Jiangsu Province, China. Ecotoxicol. Environ. Saf. 2018, 157, 169–175. [Google Scholar] [CrossRef]
- Ruiz, G.M.; Fofonoff, P.W.; Carlton, J.T.; Wonham, M.J.; Hines, A.H. Invasion of coastal marine communities in North America: Apparent patterns, processes, and biases. Annu. Rev. Ecol. Syst. 2000, 31, 481–531. [Google Scholar] [CrossRef] [Green Version]
- Davidson, I.C.; Minton, M.S.; Carney, K.J.; Miller, A.W.; Ruiz, G.M. Pioneering patterns of ballast treatment in the emerging era of marine vector management. Mar. Policy. 2017, 78, 158–162. [Google Scholar] [CrossRef]
- Drillet, G. Protect aquaculture from ship pathogens. Nature 2016, 539, 31. [Google Scholar] [CrossRef] [Green Version]
- Lavoie, D.M.; Smith, L.D.; Ruiz, G.M. The potential for intracoastal transfer of non-indigenous species in the ballast water of ships. Estuar. Coast. Shelf Sci. 1999, 48, 551–564. [Google Scholar] [CrossRef]
- Drake, L.A.; Doblin, M.A.; Dobbs, F.C. Potential microbial bioinvasions via ships’ ballast water, sediment, and biofilm. Mar. Pollut. Bull. 2007, 55, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Emami, K.; Askari, V.; Ullrich, M.; Mohinudeen, K.; Anil, A.C.; Khandeparker, L.; Burgess, J.G.; Mesbahi, E. Characterization of Bacteria in Ballast Water Using MALDI-TOF Mass Spectrometry. PLoS ONE 2012, 7, e38515. [Google Scholar] [CrossRef] [Green Version]
- Di Cesare, A.; Vignaroli, C.; Luna, G.M.; Pasquaroli, S.; Biavasco, F. Antibiotic-Resistant Enterococci in Seawater and Sediments from a Coastal Fish Farm. Microb. Drug Resist. 2012, 18, 502–509. [Google Scholar] [CrossRef]
- Akinbowale, O.L.; Peng, H.; Barton, M.D. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 2006, 100, 1103–1113. [Google Scholar] [CrossRef]
- Diehl, D.L.; LaPara, T.M. Effect of Temperature on the Fate of Genes Encoding Tetracycline Resistance and the Integrase of Class 1 Integrons within Anaerobic and Aerobic Digesters Treating Municipal Wastewater Solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef]
- Ghosh, S.; Ramsden, S.J.; LaPara, T.M. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2009, 84, 791–796. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, Y.; Pruden, A. Effect of temperature on removal of antibiotic resistance genes by anaerobic digestion of activated sludge revealed by metagenomic approach. Appl. Microbiol. Biotechnol. 2015, 99, 7771–7779. [Google Scholar] [CrossRef]
- Chen, J.; Wei, X.-D.; Liu, Y.-S.; Ying, G.-G.; Liu, S.-S.; He, L.-Y.; Su, H.-C.; Hu, L.-X.; Chen, F.-R.; Yang, Y.-Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Optimization of wetland substrates and hydraulic loading. Sci. Total Environ. 2016, 565, 240–248. [Google Scholar] [CrossRef]
- Guo, M.-T.; Yuan, Q.-B.; Yang, J. Ultraviolet reduction of erythromycin and tetracycline resistant heterotrophic bacteria and their resistance genes in municipal wastewater. Chemosphere 2013, 93, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.-B.; Guo, M.-T.; Yang, J. Fate of Antibiotic Resistant Bacteria and Genes during Wastewater Chlorination: Implication for Antibiotic Resistance Control. PLoS ONE 2015, 10, e0119403. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, S.; Le, T.-T.M.; Entenza, J.M.; Pulgarin, C. Solar photo-Fenton disinfection of 11 antibiotic-resistant bacteria (ARB) and elimination of representative AR genes. Evidence that antibiotic resistance does not imply resistance to oxidative treatment. Water Res. 2018, 143, 334–345. [Google Scholar] [CrossRef]
- Ren, S.; Boo, C.; Guo, N.; Wang, S.; Elimelech, M.; Wang, Y. Photocatalytic Reactive Ultrafiltration Membrane for Removal of Antibiotic Resistant Bacteria and Antibiotic Resistance Genes from Wastewater Effluent. Environ. Sci. Technol. 2018, 52, 8666–8673. [Google Scholar] [CrossRef] [PubMed]
- Karaolia, P.; Michael-Kordatou, I.; Hapeshi, E.; Drosou, C.; Bertakis, Y.; Christofilos, D.; Armatas, G.S.; Sygellou, L.; Schwartz, T.; Xekoukoulotakis, N.P.; et al. Removal of antibiotics, antibiotic-resistant bacteria and their associated genes by graphene-based TiO2 composite photocatalysts under solar radiation in urban wastewaters. Appl. Catal. B 2018, 224, 810–824. [Google Scholar] [CrossRef]
- Jiang, Q.; Yin, H.; Li, G.; Liu, H.; An, T.; Wong, P.K.; Zhao, H. Elimination of antibiotic-resistance bacterium and its associated/dissociative blaTEM-1 and aac(3)-II antibiotic-resistance genes in aqueous system via photoelectrocatalytic process. Water Res. 2017, 125, 219–226. [Google Scholar] [CrossRef]
- Gerhard, W.A.; Gunsch, C.K. Higher normalized concentrations of tetracycline resistance found in ballast and harbor water compared to ocean water. Mar. Pollut. Bull. 2020, 151, 110796. [Google Scholar] [CrossRef]
- Sayinli, B.; Dong, Y.J.; Park, Y.; Bhatnagar, A.; Sillanpaa, M. Recent progress and challenges facing ballast water treatment-A review. Chemosphere 2022, 291, 132776. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. Transport of harmful marine microalgae via ship’s ballast water: Management and mitigation with special reference to the Arabian Gulf region. Aquat. Ecosyst. Health Manag. 2015, 18, 290–298. [Google Scholar] [CrossRef]
- Lv, B.Y.; Cui, Y.X.; Tian, W.; Feng, D.L. Composition and influencing factors of bacterial communities in ballast tank sediments: Implications for ballast water and sediment management. Mar. Environ. Res. 2017, 132, 14–22. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Q.; Chen, J.; Wu, H. The occurrence of potential pathogenic bacteria on international ships’ ballast water at Yangshan Port, Shanghai, China. Mar. Pollut. Bull. 2022, 184, 114190. [Google Scholar] [CrossRef] [PubMed]
- Salleh, N.A.; Rosli, F.N.; Akbar, M.A.; Yusof, A.; Sahrani, F.K.; Razak, S.A.; Ahmad, A.; Usup, G.; Bunawan, H. Pathogenic hitchhiker diversity on international ships’ ballast water at West Malaysia port. Mar. Pollut. Bull. 2021, 172, 112850. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.; Goh, S.G.; Saeidi, N.; Gerhard, W.A.; Gunsch, C.K.; Gin, K.Y.H. Occurrence of Vibrio species, beta-lactam resistant Vibrio species, and indicator bacteria in ballast and port waters of a tropical harbor. Sci. Total Environ. 2018, 610, 651–656. [Google Scholar] [CrossRef]
- Rivera, I.N.G.; Souza, K.M.C.; Souza, C.P.; Lopes, R.M. Free-living and plankton-associated vibrios: Assessment in ballast water, harbor areas, and coastal ecosystems in Brazil. Front. Microbiol. 2013, 3, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobbs, F.C.; Goodrich, A.L.; Thomson, F.K.; Hynes, W. Pandemic Serotypes of Vibrio cholerae Isolated from Ships’ Ballast Tanks and Coastal Waters: Assessment of Antibiotic Resistance and Virulence Genes (tcpA and ctxA). Microb. Ecol. 2013, 65, 969–974. [Google Scholar] [CrossRef]
- Drake, L.A.; Meyer, A.E.; Forsberg, R.L.; Baier, R.E.; Doblin, M.A.; Heinemann, S.; Johnson, W.P.; Koch, M.; Rublee, P.A.; Dobbs, F.C. Potential invasion of microorganisms and pathogens via ‘interior hull fouling’: Biofilms inside ballast water tanks. Biol. Invasions 2005, 7, 969–982. [Google Scholar] [CrossRef]
- Faruque, S.M.; Albert, M.J.; Mekalanos, J.J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 1998, 62, 1301–1314. [Google Scholar] [CrossRef] [Green Version]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.B.; Sharma, V.K. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Zhao, Z.L.; Wang, J.; Han, Y.; Chen, J.W.; Liu, G.F.; Lu, H.; Yan, B.; Chen, S.S. Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 2017, 220, 909–918. [Google Scholar] [CrossRef]
- Wang, J.H.; Lu, J.; Zhang, Y.X.; Wu, J.; Luo, Y.M.; Liu, H. Metagenomic analysis of antibiotic resistance genes in coastal industrial mariculture systems. Bioresour. Technol. 2018, 253, 235–243. [Google Scholar] [CrossRef]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Editorial: Antibiotic Resistance in Aquatic Systems. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Nakanishi, S.; Tamminen, M.; Yokokawa, T.; Sato-Takabe, Y.; Ohta, K.; Chou, H.Y.; Muziasari, W.I.; Virta, M. Occurrence of sul and tet(M) genes in bacterial community in Japanese marine aquaculture environment throughout the year: Profile comparison with Taiwanese and Finnish aquaculture waters. Sci. Total Environ. 2019, 669, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Q.; Zheng, L.; Zhou, J.L.; Zhao, H. Persistence and risk of antibiotic residues and antibiotic resistance genes in major mariculture sites in Southeast China. Sci. Total Environ. 2017, 580, 1175–1184. [Google Scholar] [CrossRef]
- Gao, Q.X.; Li, Y.L.; Qi, Z.H.; Yue, Y.F.; Min, M.H.; Peng, S.M.; Shi, Z.H.; Gao, Y. Diverse and abundant antibiotic resistance genes from mariculture sites of China’s coastline. Sci. Total Environ. 2018, 630, 117–125. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Lu, J.; Wu, J.; Wang, J.H.; Lin, Y.C. Occurrence and distribution of antibiotic resistance genes in sediments in a semi-enclosed continental shelf sea. Sci. Total Environ. 2020, 720, 137712. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.G.; Zhang, K.; Zhang, Y. Occurrence and distribution of antibiotic resistance genes in the coastal area of the Bohai Bay, China. Mar. Pollut. Bull. 2016, 107, 245–250. [Google Scholar] [CrossRef]
- Na, G.S.; Zhang, W.R.; Zhou, S.Y.; Gao, H.; Lu, Z.H.; Wu, X.; Li, R.J.; Qiu, L.N.; Cai, Y.Q.; Yao, Z.W. Sulfonamide antibiotics in the Northern Yellow Sea are related to resistant bacteria: Implications for antibiotic resistance genes. Mar. Pollut. Bull. 2014, 84, 70–75. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J.; Wang, J.; Zhang, C.; Lin, Y. Occurrence and spatial distribution of antibiotic resistance genes in the Bohai Sea and Yellow Sea areas, China. Environ. Pollut. 2019, 252, 450–460. [Google Scholar] [CrossRef]
- Chen, J.Y.; Su, Z.G.; Dai, T.J.; Huang, B.; Mu, Q.L.; Zhang, Y.M.; Wen, D.H. Occurrence and distribution of antibiotic resistance genes in the sediments of the East China Sea bays. J. Environ. Sci. 2019, 81, 156–167. [Google Scholar] [CrossRef]
- Chen, B.W.; Yang, Y.; Liang, X.M.; Yu, K.; Zhang, T.; Li, X.D. Metagenomic Profiles of Antibiotic Resistance Genes (ARGs) between Human Impacted Estuary and Deep Ocean Sediments. Environ. Sci. Technol. 2013, 47, 12753–12760. [Google Scholar] [CrossRef]
- Suzuki, S.; Ogo, M.; Miller, T.W.; Shimizu, A.; Takada, H.; Siringan, M.A.T. Who possesses drug resistance genes in the aquatic environment? Sulfarnethoxazole (SMX) resistance genes among the bacterial community in water environment of Metro-Manila, Philippines. Front. Microbiol. 2013, 4, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.G.; Zhao, Y.; Li, B.; Huang, C.L.; Zhang, S.Y.; Yu, S.; Chen, Y.S.; Zhang, T.; Gillings, M.R.; Su, J.Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, K.; Wu, N.; Li, W.; Xu, W.; Zhang, Y.; Niu, Z. Estuarine sediments are key hotspots of intracellular and extracellular antibiotic resistance genes: A high-throughput analysis in Haihe Estuary in China. Environ. Int. 2020, 135, 105385. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Wu, X.M.; Yan, Q.P.; Ma, Y.; Huang, L.X.; Qin, Y.X.; Xu, X.J. Incidence of antimicrobial-resistance genes and integrons in antibiotic-resistant bacteria isolated from eels and aquaculture ponds. Dis. Aquat. Org. 2016, 120, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.C.; Hu, X.J.; Wang, L.L.; Xu, W.J.; Xu, Y.; Wen, G.L.; Li, Z.J.; Cao, Y.C. Contamination of antibiotic resistance genes (ARGs) in a typical marine aquaculture farm: Source tracking of ARGs in reared aquatic organisms. J. Environ. Sci. Health B 2020, 55, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, Y.B.; Xu, J.X.; Ling, J.Y.; Chen, J.L.; Zhou, J.L.; Zheng, L.; Du, Q.P. Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 2017, 168, 1107–1114. [Google Scholar] [CrossRef]
- Hong, B.; Ba, Y.; Niu, L.; Lou, F.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. A Comprehensive Research on Antibiotic Resistance Genes in Microbiota of Aquatic Animals. Front. Microbiol. 2018, 9, 1617. [Google Scholar] [CrossRef]
- Thiang, E.L.; Lee, C.W.; Takada, H.; Seki, K.; Takei, A.; Suzuki, S.; Wang, A.; Bong, C.W. Antibiotic residues from aquaculture farms and their ecological risks in Southeast Asia: A case study from Malaysia. Ecosyst. Health Sust. 2021, 7, 1926337. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.X.; Wu, J. Continental-scale spatio-temporal distribution of antibiotic resistance genes in coastal waters along coastline of China. Chemosphere 2020, 247, 125908. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.H.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef]
- Turgeon, P.; Michel, P.; Levallois, P.; Chevalier, P.; Daignault, D.; Crago, B.; Irwin, R.; McEwen, S.A.; Neumann, N.F.; Louie, M. Antimicrobial-resistant Escherichia coli in public beach waters in Quebec. Can. J. Occup. Ther. 2012, 23, E20–E25. [Google Scholar] [CrossRef] [Green Version]
- Kotlarska, E.; Luczkiewicz, A.; Pisowacka, M.; Burzynski, A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 2015, 22, 2018–2030. [Google Scholar] [CrossRef] [Green Version]
- Andrade, V.D.; Zampieri, B.D.; Ballesteros, E.R.; Pinto, A.B.; de Oliveira, A. Densities and antimicrobial resistance of Escherichia coli isolated from marine waters and beach sands. Environ. Monit. Assess. 2015, 187, 342. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Li, H.; Su, J.-Q. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 2015, 99, 5697–5707. [Google Scholar] [CrossRef]
- Tan, L.; Li, L.Y.; Ashbolt, N.; Wang, X.L.; Cui, Y.X.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.Q.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef]
- Cuadrat, R.R.C.; Sorokina, M.; Andrade, B.G.; Goris, T.; Davila, A.M.R. Global ocean resistome revealed: Exploring antibiotic resistance gene abundance and distribution in TARA Oceans samples. GigaScience 2020, 9, giaa046. [Google Scholar] [CrossRef]
- Eckert, E.M.; Di Cesare, A.; Stenzel, B.; Fontaneto, D.; Corno, G. Daphnia as a refuge for an antibiotic resistance gene in an experimental freshwater community. Sci. Total Environ. 2016, 571, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Ozaktas, T.; Taskin, B.; Gozen, A.G. High level multiple antibiotic resistance among fish surface associated bacterial populations in non-aquaculture freshwater environment. Water Res. 2012, 46, 6382–6390. [Google Scholar] [CrossRef]
- Jang, H.M.; Kim, Y.B.; Choi, S.; Lee, Y.; Shin, S.G.; Unno, T.; Kim, Y.M. Prevalence of antibiotic resistance genes from effluent of coastal aquaculture, South Korea. Environ. Pollut. 2018, 233, 1049–1057. [Google Scholar] [CrossRef]
- Campos, L.C.; Zahner, V.; Avelar, K.E.S.; Alves, R.M.; Pereira, D.S.G.; Brazil, J.M.V.; Freitas, F.S.; Salles, C.A.; Karaolis, D.K.R. Genetic diversity and antibiotic resistance of clinical and environmental Vibrio cholerae suggests that many serogroups are reservoirs of resistance. Epidemiol. Infect. 2004, 132, 985–992. [Google Scholar] [CrossRef]
- Liu, C.; Yoon, E.J.; Kim, D.; Shin, J.H.; Shin, J.H.; Shin, K.S.; Kim, Y.A.; Uh, Y.; Kim, H.S.; Kim, Y.R.; et al. Antimicrobial resistance in South Korea: A report from the Korean global antimicrobial resistance surveillance system (Kor-GLASS) for 2017. J. Infect. Chemother. 2019, 25, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Jiang, T.; Wei, H.; Tian, W.; Han, Y.; Chen, L.; Zhang, D.; Cui, Y. Transfer of antibiotic-resistant bacteria via ballast water with a special focus on multiple antibiotic-resistant bacteria: A survey from an inland port in the Yangtze River. Mar. Pollut. Bull. 2021, 166, 112166. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.; Amal, M.N.A.; Saad, M.Z.; Yasin, I.S.M.; Zulkiply, N.A.; Mustafa, M.; Nasruddin, N.S. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 2019, 15, 176. [Google Scholar] [CrossRef] [PubMed]
- Guidi, L.R.; Santos, F.A.; Ribeiro, A.C.S.R.; Fernandes, C.; Silva, L.H.M.; Gloria, M.B.A. Quinolones and tetracyclines in aquaculture fish by a simple and rapid LC-MS/MS method. Food Chem. 2018, 245, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Leal, J.F.; Henriques, I.S.; Correia, A.; Santos, E.B.H.; Esteves, V.I. Antibacterial activity of oxytetracycline photoproducts in marine aquaculture’s water. Environ. Pollut. 2017, 220, 644–649. [Google Scholar] [CrossRef]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef]
- Hatha, A.A.M.; Neethu, C.S.; Nikhil, S.M.; Rahiman, K.M.M.; Krishnan, K.P.; Saramma, A.V. Relatively high antibiotic resistance among heterotrophic bacteria from arctic fjord sediments than water-Evidence towards better selection pressure in the fjord sediments. Polar Sci. 2015, 9, 382–388. [Google Scholar] [CrossRef]
- Souissi, M.; Laabidi, R.; Aissa, P.; Pringault, O.; Ben Said, O. Influence of Bizerte city wastewater treatment plant (WWTP) on abundance and antibioresistance of culturable heterotrophic and fecal indicator bacteria of Bizerte Lagoon (Tunisia). Ecotoxicol. Environ. Saf. 2018, 148, 201–210. [Google Scholar] [CrossRef]
- Lv, B.; Cui, Y.; Tian, W.; Wei, H.; Chen, Q.; Liu, B.; Zhang, D.; Xie, B. Vessel transport of antibiotic resistance genes across oceans and its implications for ballast water management. Chemosphere 2020, 253, 126697. [Google Scholar] [CrossRef]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of Clinically Relevant Antibiotic Resistance Genes in Surface Water Samples Collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-p.; Yang, Y.; Lu, D.-p.; Niu, Z.-s.; Feng, J.-n.; Chen, Y.-r.; Tou, F.-y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liang, X.; Huang, X.; Zhang, T.; Li, X. Differentiating anthropogenic impacts on ARGs in the Pearl River Estuary by using suitable gene indicators. Water Res. 2013, 47, 2811–2820. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.L.; Zhu, D.; Xu, Y.Y. Co-driving factors of tidal effect on the abundance and distribution of antibiotic resistance genes in the Yongjiang Estuary, China. Environ. Res. 2022, 213, 113649. [Google Scholar] [CrossRef]
- Neshani, A.; Zare, H.; Eidgahi, M.R.A.; Kakhki, R.K.; Safdari, H.; Khaledi, A.; Ghazvini, K. LL-37: Review of antimicrobial profile against sensitive and antibiotic-resistant human bacterial pathogens. Gene Rep. 2019, 17, 100519. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, R.X.; Wei, Y.Y.; Chen, T.; Fan, J.Q.; Zhou, Z.C.; Makimilua, T.B.; Jiao, Y.N.; Chen, H. High-throughput profiling and analysis of antibiotic resistance genes in East Tiaoxi River, China. Environ. Pollut. 2017, 230, 648–654. [Google Scholar] [CrossRef]
- Prange, G.J.; Pereira, N.N. Ship Ballast Tank Sediment Reduction Methods. Nav. Eng. J. 2013, 125, 127–134. [Google Scholar]
- Casas-Monroy, O.; Roy, S.; Rochon, A. Dinoflagellate cysts in ballast sediments: Differences between Canada’s east coast, west coast and the Great Lakes. Aquat. Toxicol. 2013, 23, 254–276. [Google Scholar] [CrossRef]
- Gollasch, S.; Rosenthal, H.; Botnen, H.; Hamer, J.; Laing, I.; Leppakoski, E.; Macdonald, E.; Minchin, D.; Nauke, M.; Olenin, S.; et al. Fluctuations of zooplankton taxa in ballast water during short-term and long-term ocean-going voyages. Int. Rev. Hydrobiol. 2000, 85, 597–608. [Google Scholar] [CrossRef]
- Chen, B.; Liang, X.; Nie, X.; Huang, X.; Zou, S.; Li, X. The role of class I integrons in the dissemination of sulfonamide resistance genes in the Pearl River and Pearl River Estuary, South China. J. Hazard. Mater. 2015, 282, 61–67. [Google Scholar] [CrossRef]
- Lin, L.; Yuan, K.; Liang, X.; Chen, X.; Zhao, Z.; Yang, Y.; Zou, S.; Luan, T.; Chen, B. Occurrences and distribution of sulfonamide and tetracycline resistance genes in the Yangtze River Estuary and nearby coastal area. Mar. Pollut. Bull. 2015, 100, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xu, Y.; Chen, J.; Ling, J.; Li, Y.; Huang, L.; Zhou, X.; Zheng, L.; Xie, G. Evolution of corresponding resistance genes in the water of fish tanks with multiple stresses of antibiotics and heavy metals. Water Res. 2017, 124, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, A.Z.; He, M.; Li, D.; Chen, J. Subinhibitory Concentrations of Disinfectants Promote the Horizontal Transfer of Multidrug Resistance Genes within and across Genera. Environ. Sci. Technol. 2017, 51, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Vats, P.; Kaur, U.J.; Rishi, P. Heavy metal-induced selection and proliferation of antibiotic resistance: A review. J. Appl. Microbiol. 2022, 132, 4058–4076. [Google Scholar] [CrossRef]
- Lu, Z.; Na, G.; Gao, H.; Wang, L.; Bao, C.; Yao, Z. Fate of sulfonamide resistance genes in estuary environment and effect of anthropogenic activities. Sci. Total Environ. 2015, 527, 429–438. [Google Scholar] [CrossRef]
- Su, H.-C.; Liu, Y.-S.; Pan, C.-G.; Chen, J.; He, L.-Y.; Ying, G.-G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 2018, 616, 453–461. [Google Scholar] [CrossRef]
- Zheng, J.; Zhou, Z.; Wei, Y.; Chen, T.; Feng, W.; Chen, H. High-throughput profiling of seasonal variations of antibiotic resistance gene transport in a peri-urban river. Environ. Int. 2018, 114, 87–94. [Google Scholar] [CrossRef]
- Xu, Y.-B.; Hou, M.-Y.; Li, Y.-F.; Huang, L.; Ruan, J.-J.; Zheng, L.; Qiao, Q.-X.; Du, Q.-P. Distribution of tetracycline resistance genes and AmpC beta-lactamase genes in representative non-urban sewage plants and correlations with treatment processes and heavy metals. Chemosphere 2017, 170, 274–281. [Google Scholar] [CrossRef]
- Furukawa, T.; Hashimoto, R.; Mekata, T. Quantification of vancomycin-resistant enterococci and corresponding resistance genes in a sewage treatment plant. J. Environ. Sci. Health A 2015, 50, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512, 125–132. [Google Scholar] [CrossRef]
- McKinney, C.W.; Pruden, A. Ultraviolet Disinfection of Antibiotic Resistant Bacteria and Their Antibiotic Resistance Genes in Water and Wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, L.; Yao, S.; Lin, K.; Zhou, Y.; Cui, C. Removal of antibiotic resistance genes and control of horizontal transfer risk by UV, chlorination and UV/chlorination treatments of drinking water. Chem. Eng. J. 2019, 358, 589–597. [Google Scholar] [CrossRef]
- Zhuang, Y.; Ren, H.; Geng, J.; Zhang, Y.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater by chlorination, ultraviolet, and ozonation disinfection. Environ. Sci. Pollut. Res. 2015, 22, 7037–7044. [Google Scholar] [CrossRef] [PubMed]
- Asghari, F.B.; Dehghani, M.H.; Dehghanzadeh, R.; Farajzadeh, D.; Shanehbandi, D.; Mahvi, A.H.; Yaghmaeian, K.; Rajabi, A. Performance evaluation of ozonation for removal of antibiotic-resistant Escherichia coli and Pseudomonas aeruginosa and genes from hospital wastewater. Sci. Rep. 2021, 11, 24519. [Google Scholar] [CrossRef] [PubMed]
- Cengiz, M.; Uslu, M.O.; Balcioglu, I. Treatment of E. coli HB101 and the tetM gene by Fenton’s reagent and ozone in cow manure. J. Environ. Manag. 2010, 91, 2590–2593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Xu, K.; Ding, L. Reduction of antibiotic resistance genes in municipal wastewater effluent by advanced oxidation processes. Sci. Total Environ. 2016, 550, 184–191. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, Z.; Cheng, Z.; Zhang, G.; Li, M.; Li, Y.; Zhan, S.; Crittenden, J.C. Mechanistic insights for efficient inactivation of antibiotic resistance genes: A synergistic interfacial adsorption and photocatalytic-oxidation process. Sci. Bull. 2020, 65, 2107–2119. [Google Scholar] [CrossRef]
- Cai, Q.; Hu, J. Effect of UVA/LED/TiO2 photocatalysis treated sulfamethoxazole and trimethoprim containing wastewater on antibiotic resistance development in sequencing batch reactors. Water Res. 2018, 140, 251–260. [Google Scholar] [CrossRef]
- Li, B.; Zhang, T. Different removal behaviours of multiple trace antibiotics in municipal wastewater chlorination. Water Res. 2013, 47, 2970–2982. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, T. pH significantly affects removal of trace antibiotics in chlorination of municipal wastewater. Water Res. 2012, 46, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, D.; Du, Y. Acute toxicity evaluation for quinolone antibiotics and their chlorination disinfection processes. J. Environ. Sci. 2014, 26, 1837–1842. [Google Scholar] [CrossRef]
- Dodd, M.C. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J. Environ. Monit. 2012, 14, 1754–1771. [Google Scholar] [CrossRef]
- Furukawa, T.; Jikumaru, A.; Ueno, T.; Sei, K. Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection. Water 2017, 9, 547. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, M.; Zhang, S.; Yu, X. Can chlorination co-select antibiotic-resistance genes? Chemosphere 2016, 156, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Huang Jing, J.; Xi Jing, Y.; Hu Hong, Y.; Tang, F.; Pang Yu, C. Inactivation and Regrowth of Antibiotic-resistant Bacteria by PAA Disinfection in the Secondary Effluent of a Municipal Wastewater Treatment Plant. Biomed. Environ. Sci. 2013, 26, 865–868. [Google Scholar] [CrossRef]
- Liu, S.-S.; Qu, H.-M.; Yang, D.; Hu, H.; Liu, W.-L.; Qiu, Z.-G.; Hou, A.-M.; Guo, J.; Li, J.-W.; Shen, Z.-Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef] [Green Version]
- Serna-Galvis, E.A.; Giraldo-Aguirre, A.L.; Silva-Agredo, J.; Florez-Acosta, O.A.; Torres-Palma, R.A. Removal of antibiotic cloxacillin by means of electrochemical oxidation, TiO2 photocatalysis, and photo-Fenton processes: Analysis of degradation pathways and effect of the water matrix on the elimination of antimicrobial activity. Environ. Sci. Pollut. Res. 2017, 24, 6339–6352. [Google Scholar] [CrossRef]
- Ding, Y.; Jiang, W.; Liang, B.; Han, J.; Cheng, H.; Haider, M.R.; Wang, H.; Liu, W.; Liu, S.; Wang, A. UV photolysis as an efficient pretreatment method for antibiotics decomposition and their antibacterial activity elimination. J. Hazard. Mater. 2020, 392, 122321. [Google Scholar] [CrossRef]
- Phattarapattamawong, S.; Chareewan, N.; Polprasert, C. Comparative removal of two antibiotic resistant bacteria and genes by the simultaneous use of chlorine and UV irradiation (UV/chlorine): Influence of free radicals on gene degradation. Sci. Total Environ. 2021, 755, 142696. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.A.; Vance, C.C.; Gentry, T.J.; Karthikeyan, R. Effects of chlorination and ultraviolet light on environmental tetracycline-resistant bacteria and tet(W) in water. J. Environ. Chem. Eng. 2017, 5, 777–784. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.T.; Yuan, Q.B.; Yang, J. Distinguishing Effects of Ultraviolet Exposure and Chlorination on the Horizontal Transfer of Antibiotic Resistance Genes in Municipal Wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, M.; Zhang, K.; Wang, N.; Wang, K.; Wang, H.; Meng, S.; Mu, R. Effect of ultrasound irradiation combined with ozone pretreatment on the anaerobic digestion for the biosludge exposed to trace-level levofloxacin: Degradation, microbial community and ARGs analysis. J. Environ. Manag. 2020, 262, 110356. [Google Scholar] [CrossRef]
- Alsager, O.A.; Alnajrani, M.N.; Abuelizz, H.A.; Aldaghmani, I.A. Removal of antibiotics from water and waste milk by ozonation: Kinetics, byproducts, and antimicrobial activity. Ecotoxicol. Environ. Saf. 2018, 158, 114–122. [Google Scholar] [CrossRef]
- Zheng, S.; Cui, C.; Liang, Q.; Xia, X.; Yang, F. Ozonation performance of WWTP secondary effluent of antibiotic manufacturing wastewater. Chemosphere 2010, 81, 1159–1163. [Google Scholar] [CrossRef]
- Chen, X.; Tang, R.; Wang, Y.; Yuan, S.; Wang, W.; Ali, I.M.; Hu, Z.-H. Effect of ultrasonic and ozone pretreatment on the fate of enteric indicator bacteria and antibiotic resistance genes, and anaerobic digestion of dairy wastewater. Bioresour. Technol. 2021, 320, 124356. [Google Scholar] [CrossRef]
- Lueddeke, F.; Hess, S.; Gallert, C.; Winter, J.; Guede, H.; Loeffler, H. Removal of total and antibiotic resistant bacteria in advanced wastewater treatment by ozonation in combination with different filtering techniques. Water Res. 2015, 69, 243–251. [Google Scholar] [CrossRef]
- Alexander, J.; Knopp, G.; Doetsch, A.; Wieland, A.; Schwartz, T. Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci. Total Environ. 2016, 559, 103–112. [Google Scholar] [CrossRef]
- Pak, G.; Salcedo, D.E.; Lee, H.; Oh, J.; Maeng, S.K.; Song, K.G.; Hong, S.W.; Kim, H.-C.; Chandran, K.; Kim, S. Comparison of Antibiotic Resistance Removal Efficiencies Using Ozone Disinfection under Different pH and Suspended Solids and Humic Substance Concentrations. Environ. Sci. Technol. 2016, 50, 7590–7600. [Google Scholar] [CrossRef]
- Beretsou, V.G.; Michael-Kordatou, I.; Michael, C.; Santoro, D.; El-Halwagy, M.; Jager, T.; Besselink, H.; Schwartz, T.; Fatta-Kassinos, D. A chemical, microbiological and (eco)toxicological scheme to understand the efficiency of UV-C/H2O2 oxidation on antibiotic-related microcontaminants in treated urban wastewater. Sci. Total Environ. 2020, 744, 140835. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Chueca, J.; Varella della Giustina, S.; Rocha, J.; Fernandes, T.; Pablos, C.; Encinas, A.; Barcelo, D.; Rodriguez-Mozaz, S.; Manaia, C.M.; Marugan, J. Assessment of full-scale tertiary wastewater treatment by UV-C based-AOPs: Removal or persistence of antibiotics and antibiotic resistance genes? Sci. Total Environ. 2019, 652, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Cao, S.; Jin, W.; Zhou, X.; Ding, W.; Tu, R.; Han, S.-F.; Wang, C.; Jiang, Q.; Huang, H.; et al. Inactivation of chlorine-resistant bacterial spores in drinking water using UV irradiation, UV/Hydrogen peroxide and UV/Peroxymonosulfate: Efficiency and mechanism. J. Clean. Prod. 2020, 243, 118666. [Google Scholar] [CrossRef]
- Ferro, G.; Guarino, F.; Castiglione, S.; Rizzo, L. Antibiotic resistance spread potential in urban wastewater effluents disinfected by UV/H2O2 process. Sci. Total Environ. 2016, 560, 29–35. [Google Scholar] [CrossRef]
- Guo, C.; Wang, K.; Hou, S.; Wan, L.; Lv, J.; Zhang, Y.; Qu, X.; Chen, S.; Xu, J. H2O2 and/or TiO2 photocatalysis under UV irradiation for the removal of antibiotic resistant bacteria and their antibiotic resistance genes. J. Hazard. Mater. 2017, 323, 710–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.; Chung, H.J.; Di, D.Y.W.; Dodd, M.C.; Hur, H.-G.; Lee, Y. Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Res. 2017, 123, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Chen, Z.; Gao, J.; Xie, Y.; Li, L.; Qin, S.; Wang, Q.; Mao, D.; Luo, Y. Simultaneous removal of antibiotics and antibiotic resistance genes from pharmaceutical wastewater using the combinations of up-flow anaerobic sludge bed, anoxic-oxic tank, and advanced oxidation technologies. Water Res. 2019, 159, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Su, C.; Zhou, J.W.; Xu, L.K.; Qian, Y.Y.; Chen, H. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants. Chem. Eng. J. 2017, 317, 309–316. [Google Scholar] [CrossRef]
- Xi, C.; Zhang, Y.; Marrs, C.F.; Ye, W.; Simon, C.; Foxman, B.; Nriagu, J. Prevalence of Antibiotic Resistance in Drinking Water Treatment and Distribution Systems. Appl. Environ. Microbiol. 2009, 75, 5714–5718. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]





| Discharge Limitation Colony form Ingunit (cfu) | ||
|---|---|---|
| Indicator microbes | Toxicogenic Vibrio cholerae | <1 cfu per 100 mL |
| Escherichia coli (E.coli) | <250 cfu per 100 mL | |
| Intestinal Enterococci | <100 cfu per 100 mL | |
| Size of microorganism | ≥50 μm | <10 viable organisms per m3 |
| ≥10 μm and <50 μm | <10 viable organisms per mL |
| Category | Effect | Representative Drug |
|---|---|---|
| Tetracyclines | It is an antibiotic with a broad spectrum that destroys a variety of bacteria. Its primary function is to inhibit peptide chain growth and bacterial protein synthesis. | Tetracycline, oxytetracycline and chlortetracycline, etc. |
| Sulfonamides | It is a synthetic antibacterial drug with a broad antibacterial spectrum and stable properties. Its main function is to affect bacterial nucleoprotein synthesis by hindering the formation of dihydrofolic acid, thereby inhibiting bacterial reproduction. This kind of drug is a derivative of p-aminobenzene sulfonamide as a skeleton structure. | Sulfadiazine, Sulfathiazole, Sulfamethoxazole, etc. |
| Quinolones | It is an artificial, synthetic antibacterial agent with a broad antibacterial spectrum and strong antibacterial activity. It uses the 4-quinolone ring as the basic skeleton structure, and acts on bacterial deoxyribonucleic acid (DNA) helicase to damage chromosomes and kill bacteria. | Enrofloxacin, Norfloxacin, Ciprofloxacin, etc. |
| β-lactams | It mainly prevents the development of cell walls. It has the greatest diversity and is capable of treating most diseases. | Penicillins and cephalosporins, etc. |
| Aminoglycosides | It mainly inhibits the production of proteins by bacteria, increases the permeability of cell membranes, and causes chemicals within the cells to flow out. It removes bacteria. | Streptomycin, gentamicin, neomycin, kanamycin, etc. |
| Macrolides | It mainly inhibits bacterial protein synthesis. | Erythromycin, Tylosin, etc. |
| Amide alcohols | Broad-spectrum antibiotics produce an irreversible bond with the 50 S component of the bacterial ribosome, which then destroys microorganisms. This inhibits the formation of DNA white matter, acyl transfer, and peptide chain extension. | Chloramphenicol, Florfenicol, Thiamphenicol, etc. |
| The Marine Environment | ARGs | MGEs | Ref. |
|---|---|---|---|
| Bohai Sea | blaTEM, sul1, sul2, qnrA, tetX, qnrS | intI1 | [65,66] |
| Yellow Sea | sul1, sul2, tetG, tetX | intI1 | [67,68] |
| East China Sea | sul1, sul2, sul3, tetC, tetW, dfrA1, dfrA13, floR, blaPSE-1 | intI1 | [69] |
| South China Sea | macB, acrB, tetX, bavA, arnA | [65,70] | |
| Manila Bay | sul1, sul2, sul3 | [71] | |
| Estuary | aacC, aadA5, mphA, oprD, oprJ, qacED1, tetG, aadA1 | intI1 | [72,73] |
| Mariculture | bacA, mexF, mexB, sul1, sul2, tetA, tetB, tetC, tetD, tetW, tetQ, tetO, floR, qnrA, qnrB, qnrD, aadA, blaTEM | intI1, intI2 | [62,63,64,74,75,76,77,78,79] |
| Beaches | tetA, tetB, qnrS, sul1, dfrA5, dfrA7 | intI1 | [80,81,82,83] |
| Polar | sul1, sul2, sul3, qnrB, tetD, tetG, aadA2, qacEdlta1 | intI1, cInl1, tnpA | [84,85,86] |
| Classification | Advantage | Shortcoming | Removal Effect of ARGs | Ref. |
|---|---|---|---|---|
| Ultraviolet disinfection | No byproducts, no chemical residue, noncorrosive water treatment. | There is photoreactivation. It is difficult to achieve a higher UV dose in actual production. | At a UV dose of 5 mJ cm−2, erythromycin ARGs and tetracycline ARGs decreased by an average of 3.0 ± 0.1 log and 1.9 ± 0.1 log, respectively. UV dose of 200~400 mJ cm−2 reduced ARGs (mecA, vanA, tetA and ampC) by 3~4 log; weak effect on tetX, sul1, tetG, and intI1. At 60 min, the removal efficiency of sul1-qPCR is greater than 3.50 log, and that of intI1-qPCR is greater than 4.00 log. | [41,122,123,124] |
| Chlorination disinfection | Simple operation, low cost. | It produces toxic byproducts, causes chemical residues. | Reduce erythromycin ARGs of 0.42 ± 0.12 log and tetracycline ARGs of 0.10 ± 0.02 log. sul1, tetX, tetG, and intI1 can remove 1.30~1.49 log. When the chlorine dosage is from 5–20 mg L−1, the removal of ARGs increases slowly, and a larger free chlorine dosage will lead to higher gene removal efficiency. | [42,122,123] |
| Ozonation | Strong oxidizability, fast reaction speed, no residue. | Ozone is easy to consume. It produces harmful byproducts. | sul1 and tetG decreased by 1.65~2.28 log. Under high ozone concentration, the removal rate of blactx was slightly higher than that of qnrS gene. | [125,126] |
| Fenton oxidation | Strong oxidation, simple operation, and low cost. | Reagent consumption is high. Excessive Fe2+ will affect the effluent quality and cause secondary pollution. | With an increase in Fenton reagent concentration, the band strength of tetM gene decreased gradually. The gel electrophoresis band strength of tetM decreased with an increase in oxidant dose; under the action of Fe2+/H2O2, the concentrations of sul1, tetX, and tetG decreased by 2.58~3.79 log, while under the action of UV/H2O2, they decreased by 2.8~3.5 log. | [43,127,128] |
| Photocatalytic oxidation | Tasteless and nontoxic, strong oxidation ability and complete sterilization, mild reaction conditions. | With the progress of the reaction, the catalyst will be deactivated, and the utilization rate of the light source will not be high. | 5.8 log mecA and 4.7 log ampC were removed. The degradation efficiency of floR, tetC, sul1, and intI1 was 97.82, 20.66, 99.45, and 93.67% respectively. The synthesized graphene-based photocatalyst successfully removed ampC and significantly reduced the ecfX abundance of Pseudomonas aeruginosa. | [44,45,67,129,130] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Jiang, B.; Sumita; Wu, C.; Zhang, Y.; Li, C. Review of the Distribution and Influence of Antibiotic Resistance Genes in Ballast Water. Water 2022, 14, 3501. https://doi.org/10.3390/w14213501
Guo J, Jiang B, Sumita, Wu C, Zhang Y, Li C. Review of the Distribution and Influence of Antibiotic Resistance Genes in Ballast Water. Water. 2022; 14(21):3501. https://doi.org/10.3390/w14213501
Chicago/Turabian StyleGuo, Jiaqi, Bo Jiang, Sumita, Chengzhang Wu, Yunshu Zhang, and Cong Li. 2022. "Review of the Distribution and Influence of Antibiotic Resistance Genes in Ballast Water" Water 14, no. 21: 3501. https://doi.org/10.3390/w14213501
APA StyleGuo, J., Jiang, B., Sumita, Wu, C., Zhang, Y., & Li, C. (2022). Review of the Distribution and Influence of Antibiotic Resistance Genes in Ballast Water. Water, 14(21), 3501. https://doi.org/10.3390/w14213501