Distribution and Transfer of Antibiotic Resistance Genes in Coastal Aquatic Ecosystems of Bohai Bay
Abstract
:1. Introduction
2. Materials and Methods
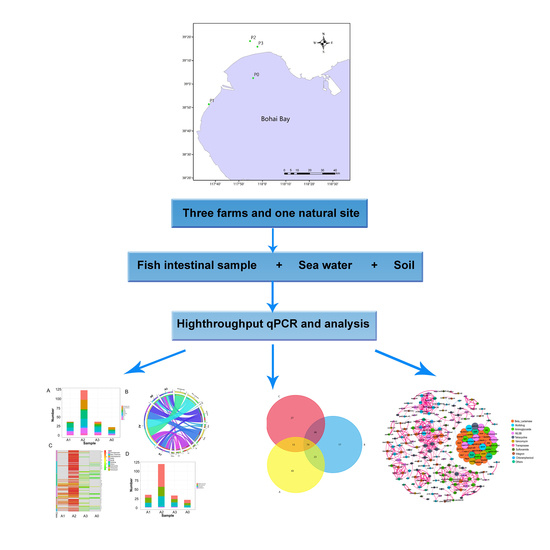
2.1. Study Sites and Sample Collection
2.2. DNA Extraction and Purification
2.3. High-Throughput Quantitative PCR
2.4. Statistical Analysis
3. Results
3.1. Distribution, Classification and Abundance of ARGs in Different Environments Associated with C. semilaevis Farming Industry
3.2. Differential Distribution of ARGs between Fish Farms and the Adjacent Natural Sea Area
3.3. Mechanisms of ARGs in Different Type of Samples
3.4. Transfer Potential of ARG Types between Fish, Soil and Water Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, H.; Zhang, J.; Chen, H.; Wang, J.; Sun, W.; Zhang, X.; Yang, Y.; Wang, Q.; Ma, J. Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci. Total Environ. 2017, 607–608, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Pontes, D.S.; de Araujo, R.S.A.; Dantas, N.; Scotti, L.; Scotti, M.T.; de Moura, R.O.; Mendonca-Junior, F.J.B. Genetic mechanisms of antibiotic resistance and the role of antibiotic adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74. [Google Scholar] [CrossRef] [PubMed]
- Loftie-Eaton, W.; Crabtree, A.; Perry, D.; Millstein, J.; Baytosh, J.; Stalder, T.; Robison, B.D.; Forney, L.J.; Top, E.M. Contagious Antibiotic Resistance: Plasmid Transfer among Bacterial Residents of the Zebrafish Gut. Appl. Environ. Microbiol. 2021, 87, e02735-20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Feng, W.Q.; Han, Y.; Zheng, J.; Chen, T.; Wei, Y.Y.; Gillings, M.; Zhu, Y.G.; Chen, H. Prevalence and transmission of antibiotic resistance and microbiota between humans and water environments. Environ. Int. 2018, 121, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Brede, D.A.; Diep, D.B.; Nes, I.F.; Lotfipour, F.; Hojabri, Z. Efficient inactivation of multi-antibiotics resistant nosocomial enterococci by purified Hiracin bacteriocin. Adv. Pharm. Bull. 2015, 5, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Muziasari, W.I.; Pitkänen, L.K.; Sørum, H.; Stedtfeld, R.D.; Tiedje, J.M.; Virta, M. The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below Baltic Sea fish farms. Front. Microbiol. 2016, 7, 2137. [Google Scholar]
- Calero-Cáceres, W.; Méndez, J.; Martín-Díaz, J.; Muniesa, M. The occurrence of antibiotic resistance genes in a Mediterranean river and their persistence in the riverbed sediment. Environ. Pollut. 2017, 223, 384–394. [Google Scholar] [CrossRef]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.L.; Andrei, A.S.; Rudi, K.; Balcázar, J.L.; Dragos, N.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 2017, 225, 304–315. [Google Scholar] [CrossRef]
- An, X.L.; Su, J.Q.; Li, B.; Ouyang, W.Y.; Zhao, Y.; Chen, Q.L.; Cui, L.; Chen, H.; Gillings, M.R.; Zhang, T.; et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environ. Int. 2018, 117, 146–153. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.H.; Qiao, M.; Su, J.Q.; Chen, Z.; Zhou, X.; Zhu, Y.G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef] [PubMed]
- Larrañaga, O.; Brown-Jaque, M.; Quirós, P.; Gómez-Gómez, C.; Blanch, A.R.; Rodríguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Muurinen, J.; Stedtfeld, R.; Karkman, A.; Pärnänen, K.; Tiedje, J.; Virta, M. Influence of manure application on the environmental resistome under finnish agricultural practice with restricted antibiotic use. Environ. Sci. Technol. 2017, 51, 5989–5999. [Google Scholar] [CrossRef]
- Pham, T.T.H.; Rossi, P.; Dinh, H.D.K.; Pham, N.T.A.; Tran, P.A.; Ho, T.; Dinh, Q.T.; De Alencastro, L.F. Analysis of antibiotic multi-resistant bacteria and resistance genes in the effluent of an intensive shrimp farm (Long An, Vietnam). J. Environ. Manag. 2018, 214, 149–156. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.X.; Zhao, Z.; Duan, C.; Chen, H.; Wang, M.; Ren, H.; Yin, Y.; Ye, L. Metagenomic analysis revealed the prevalence of antibiotic resistance genes in the gut and living environment of freshwater shrimp. J. Hazard Mater. 2018, 350, 10–18. [Google Scholar] [CrossRef]
- Almeida, A.R.; Tacão, M.; Soares, J.; Domingues, I.; Henriques, I. Tetracycline-resistant bacteria selected from water and zebrafish after antibiotic exposure. Int. J. Environ. Res. Public Health 2021, 18, 3218. [Google Scholar] [CrossRef]
- Fu, J.; Yang, D.; Jin, M.; Liu, W.; Zhao, X.; Li, C.; Zhao, T.; Wang, J.; Gao, Z.; Shen, Z.; et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017, 26, 5318–5333. [Google Scholar] [CrossRef]
- Wang, L.; Su, H.; Hu, X.; Xu, Y.; Xu, W.; Huang, X.; Li, Z.; Cao, Y.; Wen, G. Abundance and removal of antibiotic resistance genes (ARGs) in the rearing environments of intensive shrimp aquaculture in South China. J. Environ. Sci. Health B 2019, 54, 211–218. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the soil environment-degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Zhao, Y.; Li, B.; Huang, C.L.; Zhang, S.Y.; Yu, S.; Chen, Y.S.; Zhang, T.; Gillings, M.R.; Su, J.Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cui, Y.; Li, A.; Zou, X.; Ma, C.; Chen, Z. Antibiotics and antibiotic resistance genes from wastewater treated in constructed wetlands. Ecol. Eng. 2022, 177, 106548. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Melgarejo, A.; Colomer-Lluch, M.; Stoll, C.; Lucena, F.; Jofre, J.; Muniesa, M. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ. Sci. Technol. 2014, 48, 7602–7611. [Google Scholar] [CrossRef] [PubMed]
- Korzeniewska, E.; Harnisz, M. Relationship between modification of activated sludge wastewater treatment and changes in antibiotic resistance of bacteria. Sci. Total Environ. 2018, 639, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.Y.; McGrath, S.P.; Su, J.Q.; Hirsch, P.R.; Clark, I.M.; Shen, Q.; Zhu, Y.G.; Zhao, F.J. Long-term impact of field applications of sewage sludge on soil antibiotic resistome. Environ. Sci. Technol. 2016, 50, 12602–12611. [Google Scholar] [CrossRef]
- Antunes, P.; Campos, J.; Mourão, J.; Pereira, J.; Novais, C.; Peixe, L. Inflow water is a major source of trout farming contamination with Salmonella and multidrug resistant bacteria. Sci. Total Environ. 2018, 642, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhang, L.; Wang, X.; Lu, C.; Liu, J.; Liu, Y.; Li, L.; Peng, J.; Xue, M. High-throughput analysis of the effects of different fish culture methods on antibiotic resistance gene abundances in a lake. Environ. Sci. Pollut. Res. Int. 2019, 26, 5445–5453. [Google Scholar] [CrossRef]
- Yuan, J.; Ni, M.; Liu, M.; Zheng, Y.; Gu, Z. Occurrence of antibiotics and antibiotic resistance genes in a typical estuary aquaculture region of Hangzhou Bay, China. Mar. Pollut. Bull. 2019, 138, 376–384. [Google Scholar] [CrossRef]
- Ma, L.; Li, B.; Jiang, X.T.; Wang, Y.L.; Xia, Y.; Li, A.D.; Zhang, T. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome 2017, 5, 154. [Google Scholar] [CrossRef] [Green Version]
- Bello González Tde, J.; Zuidema, T.; Bor, G.; Smidt, H.; van Passel, M.W. Study of the aminoglycoside subsistence phenotype of bacteria residing in the gut of humans and zoo animals. Front. Microbiol. 2015, 6, 1550. [Google Scholar] [CrossRef] [PubMed]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of ’historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Yuan, K.; Chen, X.; Yang, Y.; Zhang, T.; Wang, Y.; Luan, T.; Zou, S.; Li, X. Metagenomic analysis revealing antibiotic resistance genes (ARGs) and their genetic compartments in the Tibetan environment. Environ. Sci. Technol. 2016, 50, 6670–6679. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; Tang, X.Y.; Cui, J.F. Effect of long-term manure slurry application on the occurrence of antibiotic resistance genes in arable purple soil (entisol). Sci. Total Environ. 2019, 647, 853–861. [Google Scholar] [CrossRef]
- Gou, M.; Hu, H.W.; Zhang, Y.J.; Wang, J.T.; Hayden, H.; Tang, Y.Q.; He, J.Z. Aerobic composting reduces antibiotic resistance genes in cattle manure and the resistome dissemination in agricultural soils. Sci. Total Environ. 2018, 612, 1300–1310. [Google Scholar] [CrossRef]
- Han, X.M.; Hu, H.W.; Shi, X.Z.; Wang, J.T.; Han, L.L.; Chen, D.; He, J.Z. Impacts of reclaimed water irrigation on soil antibiotic resistome in urban parks of Victoria, Australia. Environ. Pollut. 2016, 211, 48–57. [Google Scholar] [CrossRef]
- He, L.Y.; Liu, Y.S.; Su, H.C.; Zhao, J.L.; Liu, S.S.; Chen, J.; Liu, W.R.; Ying, G.G. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: Identification of indicator ARGs and correlations with environmental variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef]
- Kang, W.; Zhang, Y.J.; Shi, X.; He, J.Z.; Hu, H.W. Short-term copper exposure as a selection pressure for antibiotic resistance and metal resistance in an agricultural soil. Environ. Sci. Pollut. Res. Int. 2018, 25, 29314–29324. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Zhu, L.; Ge, W.; Wang, J. Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci. Total Environ. 2017, 593–594, 10–17. [Google Scholar] [CrossRef]
- Lin, H.; Chapman, S.J.; Freitag, T.E.; Kyle, C.; Ma, J.; Yang, Y.; Zhang, Z. Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers. Ecotoxicol. Environ. Saf. 2019, 170, 39–46. [Google Scholar] [CrossRef]
- Chen, Q.L.; An, X.L.; Zhu, Y.G.; Su, J.Q.; Gillings, M.R.; Ye, Z.L.; Cui, L. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 2017, 51, 8149–8157. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Chen, Q.L.; Zhu, D.; An, X.L.; Yang, X.R.; Su, J.Q.; Qiao, M.; Zhu, Y.G. Spatial and temporal distribution of antibiotic resistomes in a peri-urban area is associated significantly with anthropogenic activities. Environ. Pollut. 2018, 235, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Lin, Z.J.; Shuai, X.Y.; Zheng, J.; Meng, L.X.; Zhu, L.; Sun, Y.J.; Shang, W.C.; Chen, H. Temporal variation and sharing of antibiotic resistance genes between water and wild fish gut in a peri-urban river. J. Environ. Sci. 2021, 103, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Holmes, E.C.; Chen, X.; Tian, J.H.; Lin, X.D.; Qin, X.C.; Gao, W.H.; Liu, J.; Wu, Z.D.; Zhang, Y.Z. Diverse and abundant resistome in terrestrial and aquatic vertebrates revealed by transcriptional analysis. Sci. Rep. 2020, 10, 18870. [Google Scholar] [CrossRef]
- Nonaka, L.; Maruyama, F.; Onishi, Y.; Kobayashi, T.; Ogura, Y.; Hayashi, T.; Suzuki, S.; Masuda, M. Various pAQU plasmids possibly contribute to disseminate tetracycline resistance gene tet(M) among marine bacterial community. Front. Microbiol. 2014, 5, 152. [Google Scholar] [CrossRef]
- Chen, B.; He, R.; Yuan, K.; Chen, E.; Lin, L.; Chen, X.; Sha, S.; Zhong, J.; Lin, L.; Yang, L.; et al. Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 2017, 220, 1005–1013. [Google Scholar] [CrossRef] [Green Version]



| Site | Type | Sample | ||
|---|---|---|---|---|
| Intestinal Tracts of Fish | Sea Water | Soil | ||
| P0 | Natural sea area | A0 | B0 | C0 |
| P1 | Fish farm | A1 | B1 | C1 |
| P2 | Fish farm | A2 | B2 | C2 |
| P3 | Fish farm | A3 | B3 | C3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, L.; Liu, H.; Zhao, N.; Deng, Q.; Zhu, C.; Zhang, B. Distribution and Transfer of Antibiotic Resistance Genes in Coastal Aquatic Ecosystems of Bohai Bay. Water 2022, 14, 938. https://doi.org/10.3390/w14060938
Jia L, Liu H, Zhao N, Deng Q, Zhu C, Zhang B. Distribution and Transfer of Antibiotic Resistance Genes in Coastal Aquatic Ecosystems of Bohai Bay. Water. 2022; 14(6):938. https://doi.org/10.3390/w14060938
Chicago/Turabian StyleJia, Lei, Hao Liu, Na Zhao, Qiuxia Deng, Chunhua Zhu, and Bo Zhang. 2022. "Distribution and Transfer of Antibiotic Resistance Genes in Coastal Aquatic Ecosystems of Bohai Bay" Water 14, no. 6: 938. https://doi.org/10.3390/w14060938
APA StyleJia, L., Liu, H., Zhao, N., Deng, Q., Zhu, C., & Zhang, B. (2022). Distribution and Transfer of Antibiotic Resistance Genes in Coastal Aquatic Ecosystems of Bohai Bay. Water, 14(6), 938. https://doi.org/10.3390/w14060938