Shared 6mer Peptides of Human and Omicron (21K and 21L) at SARS-CoV-2 Mutation Sites
Abstract
:1. Introduction
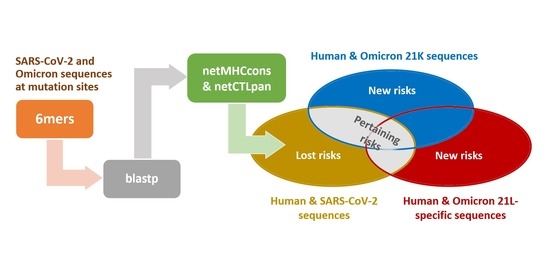
2. Materials and Methods
3. Results and Discussion
3.1. Identified Human Proteins and Peptides
3.2. Disorders, Pathways, and Expression Sites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Extended Materials and Methods
Appendix A.1.1. The Blastp Searches
Appendix A.1.2. HLA Affinity Predictions
Appendix A.1.3. Protein Features and Images
Appendix A.2. General Features of the Identified Human Proteins
- Ankyrin and Armadillo Repeat Containing (ANKAR) protein is predicted to be an integral membrane-component and the Gene Ontology annotations related to its gene include binding, binding to the nuclear receptor (Entrez, GeneCards). It is expressed in the heart and pancreatic juice (information from the estimated protein expression figure, GeneCards)
- Beta-1,3-galactosyltransferase 5 is a membrane-bound glycoprotein with galactosyltransferase and UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activities (Entrez, GeneCards).
- HGH1 homolog protein includes Maturity-Onset Diabetes of The Young, Type 3, as the associated disease (GeneCards). It is expressed in plasma, peripheral blood mononuclear cells, heart, bone, and pancreas (information retrieved from the estimated protein expression figure, GeneCards).
- HPS1 Biogenesis of Lysosomal Organelles Complex 3 Subunit 1 involves in the Hermansky-Pudlak Syndrome 1. Membrane trafficking and RAB GEF nucleotide exchange are among the pathways of its related superpathways, i.e., vesicle-mediated transport and Rab regulation of trafficking (GeneCards).
- The immunoglobulin (Ig) heavy chain variable region participates in antigen recognition, and membrane-bound immunoglobulins trigger clonal expansion and differentiation of B lymphocytes into Ig-secreting plasma cells (UniProtKB/Swiss-Prot, Entrez). Variable domains of one heavy and one (associated) light chain form two antigen binding sites with high affinity for an antigen (UniProtKB/Swiss-Prot, Entrez). Accordingly, Ig heavy chain and light chain variable regions, and the respective junction regions, are parts of the immune response.
- Mitogen-activated protein kinase kinase 3 is a dual specificity kinase, has transferase and protein tyrosine kinase activities, and its activation by cytokines, mitogens, environmental stress, and insulin is reported while the accumulation of its active form is observed during Ras oncogene expression, followed by oncogenic transformation (GeneCards, UniProtKB/Swiss-Prot, Entrez). Its inhibition is involved in the pathogenesis of Yersinia pseudotuberculosis (Entrez).
- Mucin 5AC, Oligomeric Mucus/Gel-Forming, is an extracellular matrix structural constituent, a gel-forming, protective glycoprotein of gastric and respiratory tract epithelia and interacts with H. pylori (GeneCards, UniProtKB/Swiss-Prot).
- Nucleoporin 210 is a glycoprotein and is essential for the assembly, fusion, spacing, and integrity of the nuclear pore complex, which regulates macromolecular flow (Entrez, UniProtKB/Swiss-Prot). SARS-CoV-2 infection is among the pathways in which it is involved (Superpathways, GeneCards).
- The pleckstrin homology domain containing A7 enables delta-catenin binding activity in many cellular components, resulting in epithelial cell–cell adhesion, pore complex assembly, and zonula adherens maintenance (Entrez).
- Presenilin 2 is likely a part of the catalytic subunit of the gamma-secretase complex, which is an endoprotease complex catalyzing intramembrane cleavage of integral membrane proteins (e.g., Notch receptors, amyloid-beta precursor) (UniProtKB/Swiss-Prot). It is also suggested to take part in cytoplasmic protein partitioning, intracellular signaling and gene expression, and other cellular events (UniProtKB/Swiss-Prot).
- RB Transcriptional Corepressor Like 2 (identified as 130K protein in the Blastp alignment document) is the main regulator of entry into the cell division (UniProtKB/Swiss-Prot). It “enables promoter-specific chromatin binding activity” (Entrez), can lead to (epigenetic) transcriptional repression by recruiting chromatin-modifier enzymes, histone methyltransferases, and may be involved in the transforming capacity of the adenovirus E1A protein, as well as acting as a tumor suppressor (GeneCards, UniProtKB/Swiss-Prot).
- Rho guanine nucleotide exchange factor 4 complexes with G proteins; acts as guanine nucleotide exchange factor; and stimulates Rho-dependent signals, thus participating in many extracellularly stimulated processes, as well as tumor angiogenesis (Entrez, UniProtKB/Swiss-Prot). It may play a role in intestinal adenoma formation and tumor progression (UniProtKB/Swiss-Prot).
- Ring Finger Protein 10 (identified as an unnamed protein product in the Blastp alignment document) related Gene Ontology annotations include activity of ubiquitin-protein transferase, and binding of transcription cis-regulatory region, and is involved in protein–protein interactions (GeneCards, Entrez). It is a Schwann cell differentiation and myelination regulator (UniProtKB/Swiss-Prot). Please note that the identified “unnamed protein product” had a similar sequence to the Ring Finger Protein 10 (RNF 10), although the RFN10 did not contain the region with the sequence in our results. However, the rest of its sequence was the same. Hence, the disorders, pathways, and expression sites related to RNF10 were included in the presented data.
- Signaling Lymphocytic Activation Molecule Family Member 1 is a self-ligand receptor of the signaling lymphocytic activation molecule (SLAM) family and is thus involved in modulation of the immune cell activation and differentiation, innate and adaptive immune response regulation and interconnection (UniProtKB/Swiss-Prot).
- Solute carrier family 12 member 4 (human KCC1 structure determined in KCl and detergent GDN) mediates the coupled transport of potassium and chloride ions across the plasma membrane, upon activation by the swelling of the cell (Entrez, UniProtKB/Swiss-Prot).
- Solute carrier family 22 member 6 is involved in the sodium-dependent transport and the renal elimination of endogenous and exogenous organic anions, some of which can be toxic; exchanges organic anions with a coupling; and mediates several sodium-independent uptakes (UniProtKB/Swiss-Prot, Entrez).
- Solute carrier family 25 member 27 (mitochondrial uncoupling protein 4) uncouples oxidative phosphorylation from ATP synthesis, and energy is dissipated in the form of heat as a result (UniProtKB/Swiss-Prot).
- Zinc finger protein 462 is probably involved in transcriptional regulation through the structure and organization of chromatin, leading to the regulation of, for example, pluripotency and differentiation of embryonic stem cells, and the development and differentiation of neurons (Entrez, UniProtKB/Swiss-Prot).
| Protein Disorder(s) |
| HGH1 Homolog |
| Maturity-Onset Diabetes Of The Young, Type 3 |
| Solute Carrier Family 25 Member 27 |
| Ecthyma |
| Hepatocellular Carcinoma |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Epidermolysis Bullosa Simplex 1a, Generalized Severe |
| Locked-In Syndrome |
| Beta-1,3-Galactosyltransferase 5 |
| Mood Disorder |
| Pancreatic Cancer |
| Ring Finger Protein 10 |
| Spastic Paraplegia 80, Autosomal Dominant |
| superficial keratitis |
| Ankyrin And Armadillo Repeat Containing |
| Cowden Syndrome |
| Cowden Syndrome 1 |
| Hemochromatosis, Type 4 |
| Loeys-Dietz Syndrome |
| Solute Carrier Family 22 Member 6 |
| Acute Kidney Failure |
| Fanconi Syndrome |
| Fanconi-Like Syndrome |
| Methotrexate Toxicity |
| N-Acetylglutamate Synthase Deficiency |
| Tubulointerstitial Kidney Disease, Autosomal Dominant, 1 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Breast Cancer |
| Cardiomyopathy, Familial Hypertrophic, 25 |
| Colorectal Cancer |
| Cutaneous Anthra |
| Inhalation Anthrax |
| Parkinson Disease, Late-Onset |
| Von Hippel-Lindau Syndrome |
| Solute Carrier Family 12 Member 4 |
| Agenesis Of The Corpus Callosum with Peripheral Neuropathy |
| Bartter Disease |
| Chronic Cervicitis |
| Fish-Eye Disease |
| Hemoglobin C Disease |
| Hypomagnesemia 4, Renal |
| Sickle Cell Disease |
| Pleckstrin Homology Domain Containing A7 |
| Blepharocheilodontic Syndrome 1 |
| Cleft Lip With Or Without Cleft Palate |
| Glaucoma, Primary Open Angle |
| Marshall Syndrome |
| Nanophthalmos |
| Primary Angle-Closure Glaucoma |
| Renal Adenoma |
| Stickler Syndrome |
| Zinc Finger Protein 462 |
| Acrofacial Dysostosis 1, Nager Type |
| Craniosynostosis |
| Hypermobility Syndrome |
| Metopic Ridging-Ptosis-Facial Dysmorphism Syndrome |
| Premature Menopause |
| Ptosis |
| Syndromic Intellectual Disability |
| Weiss-Kruszka Syndrome |
| Nucleoporin 210 |
| Achalasia-Addisonianism-Alacrima Syndrome |
| Amelogenesis Imperfecta, Type Ie 64 |
| Autoimmune Cholangitis |
| Autoimmune Disease Of Gastrointestinal Tract |
| Cholangitis |
| Cholangitis, Primary Sclerosing |
| Crest Syndrome |
| Peliosis Hepatis |
| Primary Biliary Cholangitis |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 |
| Albinism |
| Chediak-Higashi Syndrome |
| Hermansky-Pudlak Syndrome |
| Hermansky-Pudlak Syndrome 1 |
| Hermansky-Pudlak Syndrome Due To Bloc-3 Deficiency |
| Melanoma In Congenital Melanocytic Nevus |
| Nonspecific Interstitial Pneumonia |
| Oculocutaneous Albinism |
| Pulmonary Fibrosis |
| RB Transcriptional Corepressor Like 2 |
| Bilateral Retinoblastoma |
| Brunet-Wagner Neurodevelopmental Syndrome 6 109 |
| Burkitt Lymphoma |
| Extraocular Retinoblastoma |
| Eye Disease |
| Hypoglycemia, Leucine-Induced |
| Ocular Cancer |
| Osteogenic Sarcoma |
| Papilloma |
| Retinal Cancer |
| Retinoblastoma |
| Spastic Paraplegia 27, Autosomal Recessive |
| Spastic Paraplegia 36, Autosomal Dominant |
| Unilateral Retinoblastoma |
| Signaling Lymphocytic Activation Molecule Family Member 1 |
| Dysgammaglobulinemia |
| Herpangina |
| Immune Deficiency Disease |
| Leukemia, Acute Myeloid |
| Lymphoma, Hodgkin, Classic |
| Lymphoproliferative Syndrome 2 |
| Lymphoproliferative Syndrome |
| Lymphoproliferative Syndrome, X-Linked, 1 |
| Lymphoproliferative Syndrome, X-Linked, 2 |
| Measles |
| Pfeiffer Syndrome |
| Postinfectious Encephalitis |
| Selective Immunoglobulin Deficiency Disease |
| Subacute Sclerosing Panencephalitis |
| Systemic Lupus Erythematosus |
| Trochlear Nerve Disease |
| Viral Infectious Disease |
| Presenilin 2 |
| Acute Conjunctivitis |
| Acute Hemorrhagic Conjunctivitis |
| Agraphia |
| Alzheimer Disease 2 |
| Alzheimer Disease 3 |
| Alzheimer Disease 4 |
| Alzheimer Disease, Familial, 1 |
| Alzheimer’S Disease 1 |
| Amyloidosis |
| Amyotrophic Lateral Sclerosis 1 |
| Apperceptive Agnosia |
| Basal Ganglia Calcification |
| Breast Cancer |
| Cardiomyopathy, Dilated, 1v |
| Cerebral Amyloid Angiopathy, App-Related |
| Cerebral Amyloid Angiopathy, Cst3-Related |
| Cerebral Amyloid Angiopathy, Itm2b-Related |
| Chromosomal Disease |
| Chromosomal Duplication Syndrome |
| Conjunctival Folliculosis |
| Dementia |
| Dementia, Lewy Body |
| Dilated Cardiomyopathy |
| Disease Of Mental Health |
| Dyscalculia |
| Early-Onset Autosomal Dominant Alzheimer Disease |
| Familial Isolated Dilated Cardiomyopathy |
| Frontotemporal Dementia |
| Gerstmann Syndrome |
| Gerstmann-Straussler Disease |
| Huntington Disease-Like Syndrome |
| Hyperlucent Lung |
| Ideomotor Apraxia |
| Leber Congenital Amaurosis 7 |
| Mild Cognitive Impairment |
| Mitochondrial Dna Depletion Syndrome 12b |
| Movement Disease |
| Nervous System Disease |
| Pharyngoconjunctival Fever |
| Pick Disease Of Brain |
| Polycystic Lipomembranous Osteodysplasia With Sclerosing Leukoencephalopathy 1 |
| Posterior Cortical Atrophy |
| Prosopagnosia |
| Shipyard Eye |
| Simultanagnosia |
| Speech And Communication Disorders |
| Supranuclear Palsy, Progressive, 1 |
| Tactile Agnosia |
| Visual Agnosia |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Acute Cholangitis |
| Acute Dacryocystitis |
| Acute Inflammation Of Lacrimal Passage |
| Adenocarcinoma |
| Adenoma |
| Ampulla Of Vater Adenocarcinoma |
| Ampulla Of Vater Cancer |
| Anal Canal Adenocarcinoma |
| Anal Gland Adenocarcinoma |
| Androgen Insensitivity Syndrome |
| Anus Adenocarcinoma |
| Appendix Cancer |
| Appendix Disease |
| Asthma |
| Atopic Keratoconjunctivitis |
| Barrett Esophagus |
| Bile Duct Adenocarcinoma |
| Bile Duct Cancer |
| Bile Duct Cystadenocarcinoma |
| Bile Duct Mucinous Adenocarcinoma |
| Bile Duct Mucoepidermoid Carcinoma |
| Bile Reflux |
| Biliary Papillomatosis |
| Biliary Tract Benign Neoplasm |
| Biliary Tract Disease |
| Bladder Benign Neoplasm |
| Blepharitis |
| Breast Mucoepidermoid Carcinoma |
| Bronchial Disease |
| Bronchiolo-Alveolar Adenocarcinoma |
| Cap Polyposis |
| Cholangiocarcinoma |
| Cholecystitis |
| Chronic Asthma |
| Chronic Conjunctivitis |
| Chronic Ethmoiditis |
| Colloid Carcinoma Of The Pancreas |
| Colorectal Cancer |
| Colorectal Cancer, Hereditary Nonpolyposis, Type 8 |
| Common Cold |
| Complete Androgen Insensitivity Syndrome |
| Conjunctival Disease |
| Corneal Ulcer |
| Cystadenocarcinoma |
| Cystadenoma |
| Cystic Fibrosis |
| Cystic Teratoma |
| Dacryocystitis |
| Diverticulitis |
| Dry Eye Syndrome |
| Duodenum Adenocarcinoma |
| Duodenum Cancer |
| Duodenum Disease |
| Endobronchial Lipoma |
| Endocervical Adenocarcinoma |
| Endometrial Mucinous Adenocarcinoma |
| Exercise-Induced Bronchoconstriction |
| Eye Disease |
| Eyelid Disease |
| Filamentary Keratitis |
| Gastric Cancer |
| Gastric Tubular Adenocarcinoma |
| Inflammatory Bowel Disease |
| Interstitial Lung Disease 2 |
| Intrahepatic Biliary Papillomatosis |
| Intrahepatic Cholangiocarcinoma |
| Keratoconjunctivitis Sicca |
| Keratoconjunctivitis |
| Lacrimal Apparatus Disease |
| Limbal Stem Cell Deficiency |
| Lung Cancer Susceptibility 3 |
| Lung Disease |
| Lung Mucoepidermoid Carcinoma |
| Meconium Ileus |
| Microinvasive Gastric Cancer |
| Middle Ear Disease |
| Mucinous Adenocarcinoma |
| Mucinous Cystadenocarcinoma Of Pancreas |
| Mucinous Intrahepatic Cholangiocarcinoma |
| Mucoepidermoid Carcinoma |
| Neurotrophic Keratoconjunctivitis |
| Otitis Media |
| Ovarian Cancer |
| Ovarian Cystadenocarcinoma |
| Ovarian Mucinous Adenocarcinoma |
| Ovarian Mucinous Neoplasm |
| Pancreatic Cancer |
| Pancreatic Ductal Carcinoma |
| Pancreatic Mucinous Cystadenoma |
| Pancreatic Signet Ring Cell Adenocarcinoma |
| Poikiloderma With Neutropenia |
| Polyposis, Skin Pigmentation, Alopecia, And Fingernail Changes |
| Primary Ciliary Dyskinesia |
| Pseudomyxoma Peritonei |
| Pulmonary Disease, Chronic Obstructive |
| Punctate Epithelial Keratoconjunctivitis |
| Respiratory Allergy |
| Respiratory Failure |
| Senile Ectropion |
| Severe Cutaneous Adverse Reaction |
| Signet Ring Cell Adenocarcinoma |
| Silo Filler’S Disease |
| Small Intestine Adenocarcinoma |
| Small Intestine Cancer |
| Solid Adenocarcinoma With Mucin Production |
| Status Asthmaticus |
| T2-Low Asthma |
| Tubular Adenocarcinoma |
| Urinary Bladder Villous Adenoma |
| Vernal Conjunctivitis |
| Villous Adenoma |
| Superpathway Protein(s) |
| Jak-Stat Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| 16p11.2 Proximal Deletion Syndrome |
| Mitogen-Activated Protein Kinase Kinase 3 |
| JNK (c-Jun kinases) Phosphorylation and Activation Mediated by Activated Human TAK1 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| 4-Hydroxytamoxifen, Dexamethasone, and Retinoic Acids Regulation of p27 Expression |
| Mitogen-Activated Protein Kinase Kinase 3 |
| LKB1 Signaling Events |
| Presenilin 2 |
| ABH and Lewis Epitopes Biosynthesis from Type 1 Precursor Disaccharide |
| Beta-1,3-Galactosyltransferase 5 |
| Macrophage Differentiation and Growth Inhibition by METS |
| RB Transcriptional Corepressor Like 2 |
| Acyclovir/Ganciclovir Pathway, Pharmacokinetics/Pharmacodynamics |
| Solute Carrier Family 22 Member 6 |
| Malignant Pleural Mesothelioma |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Akt Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| MAP Kinase Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Alzheimers Disease Pathway |
| Presenilin 2 |
| MAPK Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Angiopoietin-Like Protein 8 Regulatory Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| MAPK Signaling: Oxidative Stress |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Apoptosis and Survival_Anti-Apoptotic Action of Nuclear ESR1 and ESR2 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Mesodermal Commitment Pathway |
| Zinc Finger Protein 462 |
| Zidovudine Pathway, Pharmacokinetics/Pharmacodynamics |
| Solute Carrier Family 22 Member 6 |
| Methotrexate Pathway, Pharmacokinetics |
| Solute Carrier Family 22 Member 6 |
| Bacterial Infections in CF Airways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| MicroRNAs in Cardiomyocyte Hypertrophy |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Beta-2 Adrenergic-Dependent CFTR Expression |
| Mitogen-Activated Protein Kinase Kinase 3 |
| MIF Mediated Glucocorticoid Regulation |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Blood Group Systems Biosynthesis |
| Beta-1,3-Galactosyltransferase 5 |
| Mitotic G1 Phase and G1/S Transition |
| RB Transcriptional Corepressor Like 2 |
| Canonical and Non-Canonical Notch Signaling |
| Presenilin 2 |
| Monoamine Transport |
| RB Transcriptional Corepressor Like 2 |
| Cell adhesion_Plasmin Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Nanog in Mammalian ESC Pluripotency |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Cell Cycle Regulation of G1/S Transition (Part 1) |
| RB Transcriptional Corepressor Like 2 |
| Nervous System Development |
| Presenilin 2 |
| Cellular Roles of Anthrax Toxin |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Neuropathic Pain-Signaling in Dorsal Horn Neurons |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Ceramide Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Neuroscience |
| Presenilin 2 |
| CLEC7A (Dectin-1) Signaling |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| NFAT and Cardiac Hypertrophy |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Colorectal Cancer Metastasis |
| Mitogen-Activated Protein Kinase Kinase 3 |
| NgR-p75(NTR)-Mediated Signaling |
| Rho Guanine Nucleotide Exchange Factor 4 |
| CXCR3-Mediated Signaling Events |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Non-Canonical Wnt Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Death Receptor Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Notch Pathway |
| Presenilin 2 |
| Dendritic Cells Developmental Lineage Pathway |
| Signaling Lymphocytic Activation Molecule Family Member 1 |
| Notch Signaling (Qiagen) |
| Presenilin 2 |
| Development A3 Receptor Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Notch Signaling (WikiPathways) |
| Presenilin 2 |
| Development FGFR Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Notch Signaling Pathways |
| Presenilin 2 |
| Development Notch Signaling Pathway |
| Presenilin 2 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus |
| Presenilin 2 |
| Development_TGF-beta Receptor Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| O-linked Glycosylation of Mucins |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Diseases of Glycosylation |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| p38 MAPK signaling pathway (Pathway Interaction Database) |
| Mitogen-Activated Protein Kinase Kinase 3 |
| DNA Damage |
| RB Transcriptional Corepressor Like 2 |
| P38 MAPK Signaling Pathway (sino) |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Endoderm Differentiation |
| Mitogen-Activated Protein Kinase Kinase 3 |
| p70S6K Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Epithelial to Mesenchymal Transition in Colorectal Cancer |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Phospholipase-C Pathway |
| Rho Guanine Nucleotide Exchange Factor 4 |
| FoxO Family Signaling |
| RB Transcriptional Corepressor Like 2 |
| Physiological and Pathological Hypertrophy of the Heart |
| Mitogen-Activated Protein Kinase Kinase 3 |
| G0 and Early G1 |
| RB Transcriptional Corepressor Like 2 |
| PI3K-Akt Signaling Pathway |
| RB Transcriptional Corepressor Like 2 |
| G-AlphaQ Signaling |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Pre-NOTCH Expression and Processing |
| Presenilin 2 |
| Gene Silencing by RNA |
| Nucleoporin 210 |
| Presenilin-Mediated Signaling |
| Presenilin 2 |
| Glycolysis (REACTOME) |
| Nucleoporin 210 |
| Processing of Capped Intron-Containing Pre-mRNA |
| Nucleoporin 210 |
| GPER1 Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rab Regulation of Trafficking |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 |
| G-Protein Signaling Regulation of p38 and JNK Signaling Mediated by G-proteins |
| Mitogen-Activated Protein Kinase Kinase 3 |
| RAC1 GTPase Cycle |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Guidance Cues and Growth Cone Motility |
| Rho Guanine Nucleotide Exchange Factor 4 |
| RAF/MAP Kinase Cascade |
| Presenilin 2 |
| Hematopoietic Stem Cells and Lineage-Specific Markers |
| Signaling Lymphocytic Activation Molecule Family Member 1 |
| Regulation of Actin Cytoskeleton |
| Rho Guanine Nucleotide Exchange Factor 4 |
| HIV Life Cycle |
| Nucleoporin 210 |
| Regulation of p38-alpha and p38-beta |
| Mitogen-Activated Protein Kinase Kinase 3 |
| IL-9 Signaling Pathways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Regulation of TP53 Activity |
| RB Transcriptional Corepressor Like 2 |
| Immune Response_Role of Integrins in NK Cells Cytotoxicity |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Respiratory Electron Transport, ATP Synthesis by Chemiosmotic Coupling, and Heat Production by Uncoupling Proteins |
| Solute Carrier Family 25 Member 27 |
| Influenza Infection |
| Nucleoporin 210 |
| RhoA Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Integrin-Mediated Cell Adhesion |
| Mitogen-Activated Protein Kinase Kinase 3 |
| RHOC GTPase Cycle |
| Rho Guanine Nucleotide Exchange Factor 4 |
| 4-1BB Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| RhoGDI Pathway |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Actin Nucleation by ARP-WASP Complex |
| Rho Guanine Nucleotide Exchange Factor 4 |
| SARS-CoV-2 Infection |
| Nucleoporin 210 |
| Alzheimer’s Disease and miRNA Effects |
| Presenilin 2 |
| Senescence and Autophagy in Cancer |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Antiviral Mechanism by IFN-Stimulated Genes |
| Nucleoporin 210 |
| Serotonin HTR1 Group and FOS Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| B Cell Receptor Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Sertoli-Sertoli Cell Junction Dynamics |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Beta-Adrenergic Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Signaling by ERBB4 |
| Presenilin 2 |
| CCR3 Pathway in Eosinophils |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Signaling by NOTCH3 |
| Presenilin 2 |
| Cellular Response to Heat Stress |
| Nucleoporin 210 |
| Signaling by Receptor Tyrosine Kinases |
| Presenilin 2 |
| Chromatin Regulation/Acetylation |
| RB Transcriptional Corepressor Like 2 |
| Signaling by Rho GTPases |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants |
| Presenilin 2 |
| Signaling by Slit |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Defective Binding of RB1 Mutants to E2F1,(E2F2, E2F3) |
| RB Transcriptional Corepressor Like 2 |
| Signaling Events Mediated by HDAC Class I |
| Nucleoporin 210 |
| Development Beta-Adrenergic Receptors Regulation of ERK |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Signaling Events Mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Development VEGF signaling via VEGFR2—Generic Cascades |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Signaling Events Mediated by VEGFR1 and VEGFR2 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Disorders of Transmembrane Transporters |
| Nucleoporin 210 |
| Signaling Mediated by p38-gamma and p38-delta |
| Mitogen-Activated Protein Kinase Kinase 3 |
| EPH-Ephrin Signaling |
| Presenilin 2 |
| Stabilization and Expansion of the E-cadherin Adherens Junction |
| Pleckstrin Homology Domain Containing A7 |
| FOXO-mediated Transcription |
| RB Transcriptional Corepressor Like 2 |
| Statin Pathway—Generalized, Pharmacokinetics |
| Solute Carrier Family 22 Member 6 |
| GDNF-Family Ligands and Receptor Interactions |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Sumoylation by RanBP2 Regulates Transcriptional Repression |
| Nucleoporin 210 |
| GPCR Downstream Signalling |
| Rho Guanine Nucleotide Exchange Factor 4 |
| superpathway of glycosphingolipids biosynthesis |
| Beta-1,3-Galactosyltransferase 5 |
| G-protein Signaling—Regulation of RAC1 Activity |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Sweet Taste Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| HIF1Alpha Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Tacrolimus/Cyclosporine Pathway, Pharmacodynamics |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Immune Response Fc Epsilon RI Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| TCR Signaling (Qiagen) |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Integrin Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Tenofovir/Adefovir Pathway, Pharmacokinetics |
| Solute Carrier Family 22 Member 6 |
| A-beta Plaque Formation and APP Metabolism |
| Presenilin 2 |
| Termination of O-glycan Biosynthesis |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| AMPK Enzyme Complex Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| TGF-beta Signaling Pathways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| BAFF in B-Cell Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| The Fatty Acid Cycling Model |
| Solute Carrier Family 25 Member 27 |
| Cell cycle |
| RB Transcriptional Corepressor Like 2 |
| Thermogenesis |
| Mitogen-Activated Protein Kinase Kinase 3 |
| CNTF Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| TNF Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Development A2A Receptor Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| TNF Superfamily—Human Ligand-Receptor Interactions and their Associated Functions |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Diseases Associated with O-glycosylation of Proteins |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Toll Comparative Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| fMLP Pathway |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Toll-Like receptor Signaling Pathways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Globo Sphingolipid Metabolism |
| Beta-1,3-Galactosyltransferase 5 |
| TP53 Regulates Transcription of Cell Cycle Genes |
| RB Transcriptional Corepressor Like 2 |
| HCMV Infection |
| Nucleoporin 210 |
| TRAF6 Mediated Induction of NFkB and MAP Kinases upon TLR7/8 or 9 Activation |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Inclusion Body Myositis |
| Presenilin 2 |
| Translation Insulin Regulation of Translation |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Adipogenesis |
| RB Transcriptional Corepressor Like 2 |
| Transport of Mature Transcript to Cytoplasm |
| Nucleoporin 210 |
| Breast Cancer Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Transport of the SLBP Independent Mature mRNA |
| Nucleoporin 210 |
| Cytoskeleton Remodeling Regulation of Actin Cytoskeleton by Rho GTPases |
| Presenilin 2 |
| Trk Receptor Signaling Mediated by the MAPK Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| DNA Damage Response (Only ATM Dependent) |
| RB Transcriptional Corepressor Like 2 |
| tRNA processing |
| Nucleoporin 210 |
| G-protein Signaling RAC1 in Cellular Process |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Uptake and Actions of Bacterial Toxins |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Interferon Gamma Signaling |
| Nucleoporin 210 |
| Uricosurics Pathway, Pharmacodynamics |
| Solute Carrier Family 22 Member 6 |
| Cellular Senescence |
| Mitogen-Activated Protein Kinase Kinase 3 |
| VEGF Pathway (Qiagen) |
| Mitogen-Activated Protein Kinase Kinase 3 |
| G12-G13 in Cellular Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| VEGF Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Atenolol Pathway, Pharmacokinetics |
| Solute Carrier Family 22 Member 6 |
| Vesicle-mediated Transport |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 |
| IL12-mediated Signaling Events |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Vitamin D in Inflammatory Diseases |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Development Ligand-independent Activation of ESR1 and ESR2 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Wnt/Hedgehog/Notch |
| Presenilin 2 |
| MAPK-Erk Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| RB Transcriptional Corepressor Like 2 |
| Transport of Inorganic Cations/Anions and Amino Acids/Oligopeptides |
| Solute Carrier Family 12 Member 4 |
| Solute Carrier Family 22 Member 6 |
| IL-17 Family Signaling Pathways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Cytokine Signaling in Immune System |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Nucleoporin 210 |
| TGF-Beta Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Signal Transduction |
| Presenilin 2 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Gene expression (Transcription) |
| Nucleoporin 210 |
| RB Transcriptional Corepressor Like 2 |
| Regulation of Activated PAK-2p34 by Proteasome Mediated Degradation |
| Presenilin 2 |
| RB Transcriptional Corepressor Like 2 |
| Toll-like Receptor Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Signaling Lymphocytic Activation Molecule Family Member 1 |
| Metabolism of Proteins |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Nucleoporin 210 |
| Proximal Tubule Transport |
| Solute Carrier Family 12 Member 4 |
| Solute Carrier Family 22 Member 6 |
| Glycosaminoglycan Metabolism |
| Beta-1,3-Galactosyltransferase 5 |
| Nucleoporin 210 |
| CREB Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Cell Cycle, Mitotic |
| Nucleoporin 210 |
| RB Transcriptional Corepressor Like 2 |
| Cellular Responses to Stimuli |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Nucleoporin 210 |
| GPCR Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Apoptotic Pathways in Synovial Fibroblasts |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Interferon Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Prolactin Signaling |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Presenilin 2 |
| IL-1 Family Signaling Pathways |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Thyroid Stimulating Hormone (tsh) Signaling Pathway |
| Mitogen-Activated Protein Kinase Kinase 3 |
| RB Transcriptional Corepressor Like 2 |
| p75 NTR Receptor-Mediated Signalling |
| Presenilin 2 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Innate Immune System |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Nucleoporin 210 |
| ERK Signaling |
| Presenilin 2 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Rho Guanine Nucleotide Exchange Factor 4 |
| Metabolism |
| Nucleoporin 210 |
| Beta-1,3-Galactosyltransferase 5 |
| Solute Carrier Family 25 Member 27 |
| Disease |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming |
| Nucleoporin 210 |
| Mitogen-Activated Protein Kinase Kinase 3 |
| Presenilin 2 |
| Expressing Tissue Protein(s) | Total Average Normalized Intensity Average Normalized Intensity |
| Cervical Mucosa | 3.02 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.02 |
| Osteosarcoma Cell | 3.73 |
| Solute Carrier Family 12 Member 4 | 3.73 |
| Bone | 3.85 |
| Zinc Finger Protein 462 | 3.85 |
| Oral Epithelium | 4.09 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.09 |
| Adipocyte | 4.27 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.27 |
| Uterine Cervix | 4.42 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.42 |
| Uterus | 4.67 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.67 |
| Skin | 5.31 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.31 |
| Prefrontal Cortex | 8.39 |
| Nucleoporin 210 | 3.47 |
| Solute Carrier Family 25 Member 27 | 4.92 |
| Breast | 9.12 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.77 |
| Nucleoporin 210 | 4.35 |
| Spermatozoon | 9.98 |
| Nucleoporin 210 | 5.64 |
| RB Transcriptional Corepressor Like 2 | 4.34 |
| Cardia | 11.24 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.31 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 5.93 |
| Spinal Cord | 13.50 |
| Mitogen-Activated Protein Kinase Kinase 3 | 3.99 |
| Nucleoporin 210 | 3.47 |
| Pleckstrin Homology Domain Containing A7 | 3.02 |
| Rho Guanine Nucleotide Exchange Factor 4 | 3.02 |
| Natural Killer Cell | 13.58 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.29 |
| Nucleoporin 210 | 5.41 |
| RB Transcriptional Corepressor Like 2 | 2.88 |
| Gut | 17.30 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.10 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 5.55 |
| Nucleoporin 210 | 4.58 |
| Pleckstrin Homology Domain Containing A7 | 3.07 |
| Monocyte | 17.99 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.42 |
| Nucleoporin 210 | 5.45 |
| RB Transcriptional Corepressor Like 2 | 3.62 |
| Solute Carrier Family 12 Member 4 | 3.50 |
| Pancreatic Islet | 18.45 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.85 |
| Nucleoporin 210 | 4.89 |
| Pleckstrin Homology Domain Containing A7 | 4.35 |
| Solute Carrier Family 12 Member 4 | 4.37 |
| Blood Platelet | 20.71 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 4.15 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.04 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 1.82 |
| Nucleoporin 210 | 2.14 |
| Ring Finger Protein 10 | 2.66 |
| Solute Carrier Family 12 Member 4 | 2.52 |
| Zinc Finger Protein 462 | 2.38 |
| Helper T-Lymphocyte | 22.29 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.33 |
| Nucleoporin 210 | 5.71 |
| RB Transcriptional Corepressor Like 2 | 3.67 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.33 |
| Zinc Finger Protein 462 | 3.25 |
| Heart | 22.30 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.45 |
| Nucleoporin 210 | 4.03 |
| Pleckstrin Homology Domain Containing A7 | 4.64 |
| RB Transcriptional Corepressor Like 2 | 2.84 |
| Solute Carrier Family 12 Member 4 | 3.75 |
| Solute Carrier Family 25 Member 27 | 2.60 |
| Colonic Epithelial Cell | 23.36 |
| Beta-1,3-Galactosyltransferase 5 | 5.33 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.61 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.57 |
| Nucleoporin 210 | 5.36 |
| Solute Carrier Family 25 Member 27 | 4.50 |
| Ovary | 23.41 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.14 |
| Nucleoporin 210 | 4.88 |
| Pleckstrin Homology Domain Containing A7 | 5.25 |
| RB Transcriptional Corepressor Like 2 | 3.16 |
| Solute Carrier Family 12 Member 4 | 3.73 |
| Zinc Finger Protein 462 | 2.25 |
| Urinary Bladder | 23.79 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.68 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.03 |
| Nucleoporin 210 | 3.93 |
| Pleckstrin Homology Domain Containing A7 | 4.16 |
| RB Transcriptional Corepressor Like 2 | 3.60 |
| Solute Carrier Family 12 Member 4 | 4.39 |
| B-lymphocyte | 23.82 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.49 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.74 |
| Nucleoporin 210 | 5.55 |
| RB Transcriptional Corepressor Like 2 | 4.47 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.57 |
| Cytotoxic T-lymphocyte | 24.32 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.80 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.66 |
| Nucleoporin 210 | 5.72 |
| RB Transcriptional Corepressor Like 2 | 4.47 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.66 |
| Kidney | 24.46 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.00 |
| Nucleoporin 210 | 3.89 |
| Pleckstrin Homology Domain Containing A7 | 3.83 |
| RB Transcriptional Corepressor Like 2 | 3.64 |
| Solute Carrier Family 12 Member 4 | 4.06 |
| Solute Carrier Family 22 Member 6 | 5.03 |
| Myometrium | 25.05 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 4.53 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.07 |
| RB Transcriptional Corepressor Like 2 | 3.11 |
| Ring Finger Protein 10 | 3.69 |
| Solute Carrier Family 12 Member 4 | 3.79 |
| Solute Carrier Family 25 Member 27 | 5.85 |
| Retina | 25.10 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.79 |
| Nucleoporin 210 | 4.99 |
| Pleckstrin Homology Domain Containing A7 | 4.27 |
| Solute Carrier Family 12 Member 4 | 3.17 |
| Solute Carrier Family 25 Member 27 | 4.17 |
| Zinc Finger Protein 462 | 3.70 |
| Lymph node | 25.12 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 2.76 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.21 |
| Nucleoporin 210 | 5.09 |
| RB Transcriptional Corepressor Like 2 | 4.04 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.53 |
| Solute Carrier Family 12 Member 4 | 3.49 |
| Esophagus | 27.07 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.47 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 2.89 |
| Nucleoporin 210 | 3.93 |
| Pleckstrin Homology Domain Containing A7 | 2.84 |
| RB Transcriptional Corepressor Like 2 | 4.78 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 3.38 |
| Solute Carrier Family 12 Member 4 | 4.79 |
| Liver | 27.72 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.66 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.67 |
| Nucleoporin 210 | 4.52 |
| Pleckstrin Homology Domain Containing A7 | 4.28 |
| RB Transcriptional Corepressor Like 2 | 3.66 |
| Ring Finger Protein 10 | 3.12 |
| Solute Carrier Family 12 Member 4 | 3.81 |
| Salivary Gland | 30.07 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.17 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 2.56 |
| Nucleoporin 210 | 4.00 |
| Pleckstrin Homology Domain Containing A7 | 4.48 |
| Presenilin 2 | 3.15 |
| RB Transcriptional Corepressor Like 2 | 3.38 |
| Solute Carrier Family 12 Member 4 | 4.17 |
| Zinc Finger Protein 462 | 3.17 |
| Spleen | 31.04 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.91 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 2.14 |
| Nucleoporin 210 | 4.15 |
| Pleckstrin Homology Domain Containing A7 | 2.72 |
| Presenilin 2 | 3.78 |
| RB Transcriptional Corepressor Like 2 | 4.72 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.41 |
| Solute Carrier Family 12 Member 4 | 4.19 |
| Adrenal Gland | 32.13 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.69 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.85 |
| Nucleoporin 210 | 4.26 |
| Pleckstrin Homology Domain Containing A7 | 3.41 |
| Presenilin 2 | 3.96 |
| RB Transcriptional Corepressor Like 2 | 3.86 |
| Solute Carrier Family 12 Member 4 | 4.82 |
| Zinc Finger Protein 462 | 3.28 |
| Thyroid Gland | 32.46 |
| Beta-1,3-Galactosyltransferase 5 | 3.50 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.90 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.65 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 2.53 |
| Nucleoporin 210 | 4.68 |
| Pleckstrin Homology Domain Containing A7 | 4.45 |
| RB Transcriptional Corepressor Like 2 | 3.89 |
| Solute Carrier Family 12 Member 4 | 4.85 |
| Placenta | 32.78 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.94 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.03 |
| Nucleoporin 210 | 4.48 |
| Pleckstrin Homology Domain Containing A7 | 4.47 |
| RB Transcriptional Corepressor Like 2 | 3.52 |
| Ring Finger Protein 10 | 3.46 |
| Solute Carrier Family 12 Member 4 | 5.26 |
| Zinc Finger Protein 462 | 2.63 |
| Prostate Gland | 33.04 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 4.16 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.49 |
| Nucleoporin 210 | 4.40 |
| Pleckstrin Homology Domain Containing A7 | 4.37 |
| Presenilin 2 | 4.24 |
| RB Transcriptional Corepressor Like 2 | 3.92 |
| Solute Carrier Family 12 Member 4 | 4.48 |
| Zinc Finger Protein 462 | 2.99 |
| Testis | 33.68 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.25 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.51 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 2.54 |
| Nucleoporin 210 | 4.79 |
| Pleckstrin Homology Domain Containing A7 | 3.37 |
| Presenilin 2 | 4.11 |
| RB Transcriptional Corepressor Like 2 | 3.57 |
| Solute Carrier Family 12 Member 4 | 4.19 |
| Zinc Finger Protein 462 | 3.35 |
| Stomach | 33.79 |
| Beta-1,3-Galactosyltransferase 5 | 3.44 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.02 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 6.17 |
| Nucleoporin 210 | 4.05 |
| Pleckstrin Homology Domain Containing A7 | 4.06 |
| RB Transcriptional Corepressor Like 2 | 3.11 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 3.85 |
| Solute Carrier Family 12 Member 4 | 4.09 |
| Brain | 37.57 |
| Mitogen-Activated Protein Kinase Kinase 3 | 3.85 |
| Nucleoporin 210 | 4.39 |
| Pleckstrin Homology Domain Containing A7 | 4.38 |
| Presenilin 2 | 3.24 |
| RB Transcriptional Corepressor Like 2 | 4.59 |
| Rho Guanine Nucleotide Exchange Factor 4 | 4.82 |
| Solute Carrier Family 12 Member 4 | 4.88 |
| Solute Carrier Family 25 Member 27 | 4.11 |
| Zinc Finger Protein 462 | 3.31 |
| Rectum | 37.68 |
| Beta-1,3-Galactosyltransferase 5 | 4.68 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.83 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.33 |
| Nucleoporin 210 | 4.83 |
| Pleckstrin Homology Domain Containing A7 | 3.69 |
| RB Transcriptional Corepressor Like 2 | 4.75 |
| Solute Carrier Family 12 Member 4 | 3.74 |
| Solute Carrier Family 25 Member 27 | 5.41 |
| Zinc Finger Protein 462 | 2.42 |
| Breast Cancer Cell | 37.87 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.93 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 5.99 |
| Nucleoporin 210 | 5.44 |
| Pleckstrin Homology Domain Containing A7 | 3.46 |
| Presenilin 2 | 3.46 |
| RB Transcriptional Corepressor Like 2 | 3.27 |
| Solute Carrier Family 12 Member 4 | 4.12 |
| Solute Carrier Family 22 Member 6 | 4.15 |
| Zinc Finger Protein 462 | 3.04 |
| Colon | 39.21 |
| Beta-1,3-Galactosyltransferase 5 | 4.41 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.96 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 4.38 |
| Nucleoporin 210 | 4.74 |
| Pleckstrin Homology Domain Containing A7 | 4.87 |
| RB Transcriptional Corepressor Like 2 | 4.10 |
| Solute Carrier Family 12 Member 4 | 3.99 |
| Solute Carrier Family 25 Member 27 | 4.90 |
| Zinc Finger Protein 462 | 2.87 |
| Tonsil | 39.26 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.07 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.31 |
| Nucleoporin 210 | 4.11 |
| Pleckstrin Homology Domain Containing A7 | 4.69 |
| RB Transcriptional Corepressor Like 2 | 4.31 |
| Rho Guanine Nucleotide Exchange Factor 4 | 2.74 |
| Ring Finger Protein 10 | 2.58 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.52 |
| Solute Carrier Family 12 Member 4 | 3.92 |
| Zinc Finger Protein 462 | 4.01 |
| Pancreas | 39.33 |
| Beta-1,3-Galactosyltransferase 5 | 3.69 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.31 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.64 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.80 |
| Nucleoporin 210 | 3.84 |
| Pleckstrin Homology Domain Containing A7 | 4.94 |
| Presenilin 2 | 3.86 |
| RB Transcriptional Corepressor Like 2 | 3.44 |
| Rho Guanine Nucleotide Exchange Factor 4 | 3.39 |
| Solute Carrier Family 12 Member 4 | 4.43 |
| Gall Bladder | 39.61 |
| Beta-1,3-Galactosyltransferase 5 | 4.49 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 3.52 |
| Mitogen-Activated Protein Kinase Kinase 3 | 5.08 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 5.15 |
| Nucleoporin 210 | 4.40 |
| Pleckstrin Homology Domain Containing A7 | 4.25 |
| Presenilin 2 | 3.38 |
| RB Transcriptional Corepressor Like 2 | 4.87 |
| Solute Carrier Family 12 Member 4 | 4.46 |
| Lung | 39.90 |
| HPS1 Biogenesis Of Lysosomal Organelles Complex 3 Subunit 1 | 4.18 |
| Mitogen-Activated Protein Kinase Kinase 3 | 4.48 |
| Mucin 5AC, Oligomeric Mucus/Gel-Forming | 3.98 |
| Nucleoporin 210 | 3.91 |
| Pleckstrin Homology Domain Containing A7 | 3.53 |
| Presenilin 2 | 3.85 |
| RB Transcriptional Corepressor Like 2 | 4.39 |
| Signaling Lymphocytic Activation Molecule Family Member 1 | 4.04 |
| Solute Carrier Family 12 Member 4 | 3.71 |
| Zinc Finger Protein 462 | 3.84 |
| General Sum | 1060.88 |
References
- Atyeo, C.; Fischinger, S.; Zohar, T.; Slein, M.D.; Burke, J.; Loos, C.; McCulloch, D.J.; Newman, K.L.; Wolf, C.; Yu, J.; et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 2020, 53, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, N.; Kuo, H.H.; Boucau, J.; Farmer, J.R.; Allard-Chamard, H.; Mahajan, V.S.; Piechocka-Trocha, A.; Lefteri, K.; Osborn, M.; Bals, J.; et al. The Massachusetts Consortium on Pathogen Readiness. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 2020, 183, 143–157.e13. [Google Scholar] [CrossRef] [PubMed]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.G.; Lorenc, A.; del Molino del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA genotypes, AB0 blood type and chemokine receptor 5 mutant CD195 with the clinical course of COVID-19. Eur. J. Med. Res. 2021, 1, 107. [Google Scholar] [CrossRef]
- Weiner, J.; Suwalski, P.; Holtgrewe, M.; Rakitko, A.; Thibeault, C.; Müller, M.; Patriki, D.; Quedenau, C.; Krüger, U.; Ilinsky, V.; et al. Increased risk of severe clinical course of COVID-19 in carriers of HLA-C*04:01. E Clin. Med. 2021, 40, 101099. [Google Scholar] [CrossRef]
- Gutiérrez-Bautista, J.F.; Rodriguez-Nicolas, A.; Rosales-Castillo, A.; López-Ruz, M.Á.; Martín-Casares, A.M.; Fernández-Rubiales, A.; Anderson, P.; Garrido, F.; Ruiz-Cabello, F.; López-Nevot, M.Á. Study of HLA-A; -B; -C; -DRB1 and -DQB1 polymorphisms in COVID-19 patients. J. Microbiol. Immunol. Infect 2021, S1684–S1182, 00183. [Google Scholar] [CrossRef]
- Alnaqbi, H.; Tay, G.K.; Jelinek, H.F.; Francis, A.; Alefishat, E.; El Haj Chehadeh, S.; Tahir Saeed, A.; Hussein, M.; Salameh, L.; Mahboub, B.H.; et al. HLA repertoire of 115 UAE nationals infected with SARS-CoV-2. Hum. Immunol. 2021, S0198–S8859, 00211. [Google Scholar] [CrossRef]
- Woodruff, M.C.; Ramonell, R.P.; Cashman, K.S.; Nguyen, D.C.; Saini, A.S.; Haddad, N.; Ley, A.M.; Kyu, S.; Howell, J.C.; Ozturk, T.; et al. Dominant extrafollicular B cell responses in severe COVID-19 disease correlate with robust viral-specific antibody production but poor clinical outcomes. MedRxiv 2020. [Google Scholar] [CrossRef]
- Rodríguez, Y.; Novelli, L.; Rojas, M.; De Santis, M.; Acosta-Ampudia, Y.; Monsalve, D.M.; Ramírez-Santana, C.; Costanzo, A.; Ridgway, W.M.; Ansari, A.A.; et al. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J. Autoimmun. 2020, 114, 102506. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, E.; Bramanti, A.; Ciurleo, R.; Tchorbanov, A.I.; Giordano, A.; Fagone, P.; Belizna, C.; Bramanti, P.; Shoenfeld, Y.; Nicoletti, F. Entangling COVID-19 associated thrombosis into a secondary antiphospholipid antibody syndrome: Diagnostic and therapeutic perspectives (Review). Int. J. Mol. Med. 2020, 46, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Lucchese, G. Cerebrospinal fluid findings in COVID-19 indicate autoimmunity. Lancet Microbe 2020, 1, e242. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Beziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Uppal, N.N.; Kello, N.; Shah, H.H.; Khanin, Y.; De Oleo, I.R.; Epstein, E.; Sharma, P.; Larsen, C.P.; Bijol, V.; Jhaveri, K.D. De novo ANCA-associated vasculitis with glomerulonephritis in COVID-19. Kidney Int. Rep. 2020, 5, 2079–2083. [Google Scholar] [CrossRef]
- Ryabkova, V.A.; Churilov, L.P.; Shoenfeld, Y. Influenza infection; SARS; MERS and COVID-19: Cytokine storm-The common denominator and the lessons to be learned. Clin. Immunol. 2021, 223, 108652. [Google Scholar] [CrossRef]
- David, P.; Shoenfeld, Y. The smell in COVID-19 infection: Diagnostic opportunities. Isr. Med. Assoc. J. 2020, 22, 401–403. [Google Scholar]
- Cavalli, E.; Petralia, M.C.; Basile, M.S.; Bramanti, A.; Bramanti, P.; Nicoletti, F.; Spandidos, D.A.; Shoenfeld, Y.; Fagone, P. Transcriptomic analysis of COVID19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int. J. Mol. Med. 2020, 46, 1266–1273. [Google Scholar]
- Dotan, A.; Muller, S.; Kanduc, D.; David, P.; Halpert, G.; Shoenfeld, Y. The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun. Rev. 2021, 20, 102792. [Google Scholar] [CrossRef]
- Ehrenfeld, M.; Tincani, A.; Andreoli, L.; Cattalini, M.; Greenbaum, A.; Kanduc, D.; Alijotas-Reig, J.; Zinserling, V.; Semenova, N.; Amitah, H.; et al. Covid-19 and autoimmunity. Autoimmun. Rev. 2020, 8, 102597. [Google Scholar] [CrossRef] [PubMed]
- Halpert, G.; Shoenfeld, Y. SARS-CoV-2, the autoimmune virus. Autoimmun. Rev. 2020, 19, 102695. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Rodríguez, Y.; Monsalve, D.M.; Acosta-Ampudia, Y.; Camacho, B.; Gallo, J.E.; Rojas-Villarraga, A.; Ramírez-Santana, C.; Díaz-Coronado, J.C.; Manrique, R.; et al. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020, 19, 102554. [Google Scholar] [CrossRef] [PubMed]
- Perricone, C.; Bartoloni, E.; Bursi, R.; Cafaro, G.; Guidelli, G.M.; Shoenfeld, Y.; Gerli, R. COVID-19 as part of the hyperferritinemic syndromes: The role of iron depletion therapy. Immunol. Res. 2020, 68, 213–224. [Google Scholar] [CrossRef]
- Shoenfeld, Y. Corona (COVID-19) time musings: Our involvement in COVID-19 pathogenesis; diagnosis; treatment and vaccine planning. Autoimmun. Rev. 2020, 19, 102538. [Google Scholar] [CrossRef]
- Kanduc, D.; Shoenfeld, Y. On the molecular determinants of the SARS-CoV-2 attack. Clin. Immunol. 2020, 215, 108426. [Google Scholar] [CrossRef]
- Kanduc, D.; Shoenfeld, Y. Medical, genomic, and evolutionary aspects of the peptide sharing between pathogens, primates, and humans. Global Med. Genet. 2020, 7, 64–67. [Google Scholar] [CrossRef]
- Kanduc, D.; Shoenfeld, Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: Implications for the vaccine. Immunol. Res. 2020, 68, 310–313. [Google Scholar] [CrossRef]
- Kanduc, D. From anti-SARS-CoV-2 immune responses to COVID-19 via molecular mimicry. Antibodies 2020, 9, 33. [Google Scholar] [CrossRef]
- Lucchese, G.; Flöel, A. Molecular mimicry between SARS-CoV-2 and respiratory pacemaker neurons. Autoimmun. Rev. 2020, 19, 102556. [Google Scholar] [CrossRef]
- Lucchese, G.; Flöel, A. SARS-CoV-2 and Guillain-Barré syndrome: Molecular mimicry with human heat shock proteins as potential pathogenic mechanism. Cell Stress Chaperones 2020, 25, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Angileri, F.; Legare, S.; Gammazza, A.M.; de Macario, E.C.; Macario, A.J.; Cappello, F. Molecular mimicry may explain multi-organ damage in COVID-19. Autoimmun. Rev. 2020, 19, 102591. [Google Scholar] [CrossRef] [PubMed]
- Vojdani, A.; Kharrazian, D. Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune diseases. Clin. Immunol. 2020, 217, 108480. [Google Scholar] [CrossRef] [PubMed]
- Lyons-Weiler, J. Pathogenic priming likely contributes to serious and critical illness and mortality in COVID-19 via autoimmunity. J. Transl. Autoimmun. 2020, 3, 100051. [Google Scholar] [CrossRef] [PubMed]
- Adiguzel, Y. Molecular mimicry between SARS-CoV-2 and human proteins. Autoimmun. Rev. 2021, 20, 102791. [Google Scholar] [CrossRef]
- Adıgüzel, Y. Molecular mimicry with Nsp11 protein of SARS-CoV-2 in individuals with HLA-B*15:01 allele. Turk. J. Immunol. 2021, 9, 95–104. [Google Scholar] [CrossRef]
- Adiguzel, Y.; Shoenfeld, Y. In Silico Study on Molecular Mimicry Based Autoimmunity Sourced by Omicron the Variant. In Proceedings of the 13th International Congress on Autoimmunity, Athens, Greece, 10–13 June 2022. [Google Scholar]
- An, H.; Park, J. Molecular mimicry map (3M) of SARS-CoV-2: Prediction of potentially immunopathogenic SARS-CoV-2 epitopes via a novel immunoinformatic approach. BioRxiv 2020. [Google Scholar] [CrossRef]
- Matzaraki, V.; Kumar, V.; Wijmenga, C.; Zhernakova, A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017, 18, 76. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Chen, Q.; Chen, J.; Chen, K.; Shen, X.; Jian, H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005, 579, 2623–2628. [Google Scholar] [CrossRef] [Green Version]
- Pavel, A.; del Giudice, G.; Federico, A.; Di Lieto, A.; Kinaret, P.A.S.; Serra, A.; Greco, D. Integrated network analysis reveals new genes suggesting COVID-19 chronic effects and treatment. Brief Bioinform. 2021, 22, 1430–1441. [Google Scholar] [CrossRef]
- Geyer, P.E.; Arend, F.M.; Doll, S.; Louiset, M.-L.; Winter, S.V.; Müller-Reif, J.B.; Torun, F.M.; Weigand, M.; Eichhorn, P.; Bruegel, M.; et al. High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion. EMBO Mol. Med. 2021, 13, e14167. [Google Scholar] [CrossRef] [PubMed]
- Sharif-Askari, N.S.; Sharif-Askari, F.S.; Ahmed, S.B.M.; Hannawi, S.; Hamoudi, R.; Hamid, Q.; Halwani, R. Enhanced expression of autoantigens during SARS-CoV-2 viral infection. Front. Immunol. 2021, 12, 2271. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, M.; Aldunate, F.; Arce, R.; Ferreiro, I.; Cristina, J. An evolutionary insight into Severe Acute Respiratory Syndrome Coronavirus 2 Omicron variant of concern. Virus Res. 2022, 314, 198753. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Dhawan, M.; Nafady, M.H.; Emran, T.B.; Mitra, S.; Choudhary, O.P.; Akter, A. Understanding the omicron variant (B.1.1.529) of SARS-CoV-2: Mutational impacts, concerns, and the possible solutions. Ann. Med. Surg 2022, 78, 103737. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; Scola, B.L.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef]
- Mungmunpuntipantip, R.; Wiwanitkit, V. Pattern of molecular mimicry between spike protein of SARS CoV2 and human thrombopoietin in beta, delta and omicron variants: A basic pathophysiological process of COVID-19 related thrombocytopenia. Am. J. Blood Res. 2022, 12, 60–63. [Google Scholar]
- An, H.; Eun, M.; Yi, J.; Park, J. CRESSP: A comprehensive pipeline for prediction of immunopathogenic SARS-CoV-2 epitopes using structural properties of proteins. Brief. Bioinform. 2022, 23, bbac056. [Google Scholar] [CrossRef]
- Nunez-Castilla, J.; Stebliankin, V.; Baral, P.; Balbin, C.A.; Sobhan, M.; Cickovski, T.; Mondal, A.M.; Narasimhan, G.; Chapagain, P.; Mathee, K.; et al. Potential autoimmunity resulting from molecular mimicry between SARS-CoV-2 Spike and human proteins. BioRxiv 2022. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022, 126, 102779. [Google Scholar] [CrossRef]
- Ameratunga, R.; Leung, E.; Woon, S.-T.; Chan, L.; Steele, R.; Lehnert, K.; Longhurst, H. SARS-CoV-2 omicron: Light at the end of the long pandemic tunnel or another false dawn for immunodeficient patients? J. Allergy Clin. Immunol. 2022, 10, 2267–2273. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, O.; Doss, A.M.; Kaplonek, P.; Liang, C.-Y.; VanBlargan, L.A.; Chen, R.E.; Monroy, J.M.; Wedner, H.J.; Kulczycki, A., Jr.; Mantia, T.L.; et al. mRNA vaccine boosting enhances antibody responses against SARS-CoV-2 Omicron variant in individuals with antibody deficiency syndromes. Cell Rep. Med. 2022, 3, 100653. [Google Scholar] [CrossRef] [PubMed]
- Peled, Y.; Afek, A.; Kreiss, Y.; Rahav, G.; Nemet, I.; Kliker, L.; Indenbaum, V.; Ram, E.; Lavee, J.; Segev, A.; et al. Kinetics of cellular and humoral responses to third BNT162B2 COVID-19 vaccine over six months in heart transplant recipients−implications for the omicron variant. J. Heart Lung Transpl. 2022, 41, 1417–1425. [Google Scholar] [CrossRef]
- Schwarze, M.; Krizsan, A.; Brakel, A.; Pohl, F.; Volke, D.; Hoffman, R. Cross-reactivity of IgG antibodies and virus neutralization in mRNA-vaccinated people against wild-type SARS-CoV-2 and the five most common SARS-CoV-2 variants of concern. Front. Immunol. 2022, 13, 915034. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 46, D8–D13. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Karosiene, E.; Lundegaard, C.; Lund, O.; Nielsen, M. NetMHCcons: A consensus method for the major histocompatibility complex class I predictions. Immunogenetics 2012, 64, 177–186. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.; Lundegaard, C.; Worning, P.; Lauemoller, S.L.; Lamberth, K.; Buus, S.; Brunak, S.; Lund, O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein. Sci. 2003, 12, 1007–1017. [Google Scholar] [CrossRef]
- Zhang, H.; Lund, O.; Nielsen, M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: Application to MHC-peptide binding. Bioinformatics 2009, 25, 1293–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Stranzl, T.; Larsen, M.V.; Lundegaard, C.; Nielsen, M. NetCTLpan. Pan-specific MHC class I pathway epitope predictions. Immunogenetics 2020, 62, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef]
- Rappaport, S.; Fishilevich, S.; Nudel, R.; Twik, M.; Belinky, F.; Plaschkes, I.; Iny Stein, T.; Cohen, D.; Oz-Levi, D.; Safran, M.; et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. BioMed. Eng. OnLine 2017, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [Green Version]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-source consolidation of human biological pathways. Database 2015, 2015, bav006. [Google Scholar] [CrossRef]
- Samaras, P.; Schmidt, T.; Frejno, M.; Gessulat, S.; Reinecke, M.; Jarzab, A.; Zecha, J.; Mergner, J.; Giansanti, P.; Ehrlich, H.-C.; et al. ProteomicsDB: A multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2020, 48, D1153–D1163. [Google Scholar] [CrossRef]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.-C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 46, D1271–D1281. [Google Scholar] [CrossRef] [Green Version]
- Stelzer, G.; Rosen, R.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny, S.T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards suite: From gene data mining to disease genome sequence analysis. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lai, Y.-C.; Cheng, Y.-W.; Chao, C.-H.; Chang, Y.-Y.; Chen, C.-D.; Tsai, W.-j.; Wang, S.; Lin, Y.-S.; Chang, C.-P.; Chuang, W.-J.; et al. Antigenic cross-reactivity between SARS-CoV-2 S1-RBD and its receptor ACE2. Front. Immunol. 2022, 13, 868724. [Google Scholar] [CrossRef] [PubMed]
- Mengist, H.M.; Kombe, A.J.K.; Mekonnen, D.; Abebaw, A.; Getachew, M.; Jin, T. Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity. Sem. Immunol. 2021, 55, 101533. [Google Scholar] [CrossRef]
- Araf, Y.; Akter, F.; Tang, Y.-D.; Fatemi, R.; Parvez, S.A.; Zheng, C.; Hossain, G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022, 94, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Farheen, S.; Araf, Y.; Tang, Y.-D.; Zheng, C. The Deltacron conundrum: Its origin and potential health risks. J. Med. Virol. 2022, 94, 5096–5102. [Google Scholar] [CrossRef]
- Adiguzel, Y.; Shoenfeld, Y. Adiguzel-Shoenfeld_dataset2. Mendeley Data 2022. [Google Scholar] [CrossRef]









| Omicron Peptide | Human Peptide | Human Protein Name | Human Protein ID | Allele | Tool for Prediction |
|---|---|---|---|---|---|
| NLAPFFTF | LLSPFFTF | Ig kappa chain variable region | ABA71433.1 | HLA-A*24:02 | NetCTLpan |
| NLAPFFTF | LLSPFFTF | Ig kappa chain variable region | ABA71433.2 | HLA-B*15:01 | NetCTLpan |
| NLAPFFTF | YLSPFFTY | hCG2003071 | EAW54993.1 | HLA-B*15:01 | NetCTLpan |
| YNLAPFFTF | YYLSPFFTY | hCG2003071 | EAW54993.1 | HLA-A*24:02 | NetCTLpan/NetMHCcons |
| NFAPF-FAF | FAPFLFAF | hCG2023603 | EAW76558.1 | HLA-A*24:02 | NetCTLpan |
| FPLRSYSF | FPLRSFSY | Ig heavy chain junction region | MOM40044.1 | HLA-B*07:02 | NetCTLpan |
| HLA 1 | SARS-CoV-2 Peptide | Prediction 2 | Human Peptide | Prediction 2 | Human Protein Name | Human Protein ID 3 | |
| 1 | A2 | TLACFVLA | WB | TLACFVAI | WB | Presenilin 2 (Alzheimer disease 4), isoform CRA_b | EAW69797.1 |
| 2 | FLACFVLV | SB | Adenosine receptor A2b | NP_000667.1 | |||
| 3 | B7 | SPRRARSV | SB/E | SPRRARII | SB | Zinc finger protein 462 isoform X1 | XP_006717272.1 |
| 4 | SPRRARGH | WB | Pleckstrin homology domain-containing family A member 7 isoform X1 | XP_047282382.1 | |||
| 5 | GPRRARSA | WB | Unnamed protein product 4 | BAG54301.1 | |||
| 6 | PPRRARSV | WB | RhoGEF | AAF79955.1 | |||
| 7 | SPRRARSS | WB | Hermansky-Pudlak syndrome 1, isoform CRA_c | EAW49879.1 | |||
| 8 | B7 | PPTSFGPL | WB | VPTSFGPL | SB | hCG1989297, isoform CRA_a | EAW55845.1 |
| 9 | B8 | SPRRARSV | WB/E | SPRRARII | WB | Zinc finger protein 462 isoform X1 | XP_006717272.1 |
| 10 | B44 | SEETGTLI | WB/E | PETGTLIV | WB | hCG1656811 | EAW75628.1 |
| 11 | B44 | EETGTLIV | WB | ||||
| HLA 1 | Omicron Peptide | Prediction 2 | Human Peptide | Prediction 2 | Protein Name | Human Protein ID 3 | |
| 12 | A1 | SGNYNYLY | WB/E | GLSGNYNY | WB | Immunoglobulin heavy chain junction region | MOL95178.1 |
| 13 | YGSGNYNY | WB | Immunoglobulin heavy chain junction region | MOL73314.1 | |||
| 14 | GSGNYNYY | SB | Immunoglobulin heavy chain junction region | MBB1884951.1 | |||
| 15 | SGNYNYFY | WB | Immunoglobulin heavy chain junction region | MOL21912.1 | |||
| 16 | A1 | LTSFGPLV | WB | ILTSFGPY | WB | Dual specificity mitogen-activated protein kinase kinase 3 isoform X1 | XP_016880346.2 |
| 17 | B39 | MHSALRLV | WB | DRHSALRL | WB | Human KCC1 structure determined in KCl and detergent GDN | 6KKR_A |
| 18 | B44 | SEEIGTLI | WB/E | AEEEIGTL | SB | 130K protein 5 | CAA53661.1 |
| 19 | B44 | EEIGTLIV | WB/E | ||||
| 20 | B62 | FLARGVVF | SB/E | AGARGVVF | WB | Immunoglobulin light chain junction region | MCC96497.1 |
| 21 | SGARGVVF | WB | Immunoglobulin light chain junction region | MCB29717.1 |
| HLA 1 | SARS-CoV-2 Peptide | Prediction 2 | Human Peptide | Prediction 2 | Human Protein Name | Human Protein ID 3 | |
| 1 | A1 | RTQLPPAY | WB/E | SIQLPPAY | E | Immunoglobulin light chain junction region | MCD11024.1 |
| 2 | A3 | FLGVYYHK | WB/E | GTFLGVYY | WB | Immunoglobulin heavy chain junction region | MBN4196023.1 |
| 3 | A3 | VLLPLTQY | WB | RLLPLTQY | WB | Protein HGH1 homolog | NP_057542.2 |
| 4 | RLLPLTQR | WB | Mitochondrial uncoupling protein 4 isoform 1 | NP_004268.3 | |||
| 5 | VLLPLTYY | WB | Immunoglobulin heavy chain junction region | MBN4485217.1 | |||
| 6 | KVLLPLTY | WB | Signaling lymphocytic activation molecule isoform a precursor | NP_001317683.1 | |||
| 7 | A26 | NSASFSTF | E | SVASFSTF | SB | Immunoglobulin heavy chain variable region, partial | UNJ97266.1 |
| 8 | B58 | RTQLPPAY | E | IQLPPAYW | SB | Immunoglobulin heavy chain junction region | MOQ03906.1 |
| 9 | B58 | NSASFSTF | WB/E | ASFSTFTI | WB | Immunoglobulin heavy chain variable region, partial | UNJ97266.1 |
| 10 | B62 | RTQLPPAY | WB | YQLPPAYY | WB | Immunoglobulin heavy chain junction region | MCG70934.1 |
| 11 | CQLPPAYY | WB | Ankyrin and armadillo repeat-containing protein isoform X1 | XP_011508975.1 | |||
| 12 | B62 | VLYNSASF | SB/E | YNSASFTF | WB | Immunoglobulin light chain junction region | MBB1719028.1 |
| 13 | B62 | NSASFSTF | WB/E | SVASFSTF | SB | Immunoglobulin heavy chain variable region, partial | UNJ97266.1 |
| 14 | B62 | KGAGGHSY | WB | QGAGGHSY | WB | Immunoglobulin heavy chain junction region | MBN4552893.1 |
| 15 | B62 | VLLPLTQY | WB | VLLPLTYY | WB | Immunoglobulin heavy chain junction region | MBN4485217.1 |
| 16 | RLLPLTQY | WB | Protein HGH1 homolog | NP_057542.2 | |||
| HLA 1 | Omicron 21L Peptide | Prediction 2 | Human Peptide | Prediction 2 | Human Protein Name | Human Protein ID 3 | |
| 17 | A1 | FLDVYYHK | WB | FLDVYYGM | WB | Immunoglobulin heavy chain junction region | MBN4448374.1 |
| 18 | FLDVYYYY | SB | Immunoglobulin heavy chain junction region | MCG72449.1 | |||
| 19 | FLDVYYNL | WB | Beta-1,3-galactosyltransferase 5 isoform b | NP_149362.2 | |||
| 20 | A3 | VLLPFTQY | WB/E | KVLLPFTR | WB | Nuclear pore membrane glycoprotein 210 precursor | NP_079199.2 |
| 21 | A24 | DYSVLYNF | WB/E | SQSVLYNF | WB | Immunoglobulin light chain variable region, partial | AHZ09416.1 |
| 22 | A24 | LYNFAPFF | SB/E | YNFAPFTF | WB | Immunoglobulin light chain junction region | MCE34472.1 |
| 23 | A24 | NFAPFFAF | SB/E | VSAPFFAF | WB | Solute carrier family 22 member 6 isoform a | NP_004781.2 |
| 24 | SAPFFAFF | WB | Solute carrier family 22 member 6 isoform a | NP_004781.2 | |||
| 25 | B7 | FPLRSYGF | WB/E | SPLRSYGM | WB | Immunoglobulin heavy chain junction region | MBB2034746.1 |
| 26 | B27 | HRYGADLK | SB/E | HRYGADYY | WB | Immunoglobulin heavy chain junction region | MBB1980753.1 |
| 27 | B27 | ARLCAKHY | WB/E | LRARLCAK | SB | Mucin, partial | AAC15950.1 |
| 28 | ARLCAKGV | WB | Mucin, partial | AAC15950.1 | |||
| 29 | B62 | VLYNFAPF | SB/E | YNFAPFTF | WB | Immunoglobulin light chain junction region | MCE34472.1 |
| SARS-CoV-2 Similar Sequences at Omicron (21K + 21K/21L) Sites | Similar of Omicron (21K + 21K/21L) Sequences with Mutations | SARS-CoV-2 Similar Sequences at Omicron 21L Sites | Similar of Sequences with Omicron 21L-Specific Mutations | |||||
|---|---|---|---|---|---|---|---|---|
| WB | SB | WB | SB | WB | SB | WB | SB | |
| Orf1ab | 0 | 1 | 3 | 0 | 7 1 | 0 | 3 | 1 |
| Spike | 5 | 1 | 3 | 1 | 6 | 3 | 8 | 1 |
| Orf9b | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Envelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Matrix | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 7 | 3 | 7 | 2 | 13 | 3 | 11 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adiguzel, Y.; Shoenfeld, Y. Shared 6mer Peptides of Human and Omicron (21K and 21L) at SARS-CoV-2 Mutation Sites. Antibodies 2022, 11, 68. https://doi.org/10.3390/antib11040068
Adiguzel Y, Shoenfeld Y. Shared 6mer Peptides of Human and Omicron (21K and 21L) at SARS-CoV-2 Mutation Sites. Antibodies. 2022; 11(4):68. https://doi.org/10.3390/antib11040068
Chicago/Turabian StyleAdiguzel, Yekbun, and Yehuda Shoenfeld. 2022. "Shared 6mer Peptides of Human and Omicron (21K and 21L) at SARS-CoV-2 Mutation Sites" Antibodies 11, no. 4: 68. https://doi.org/10.3390/antib11040068
APA StyleAdiguzel, Y., & Shoenfeld, Y. (2022). Shared 6mer Peptides of Human and Omicron (21K and 21L) at SARS-CoV-2 Mutation Sites. Antibodies, 11(4), 68. https://doi.org/10.3390/antib11040068