Precision Medicine and Public Health: New Challenges for Effective and Sustainable Health
Abstract
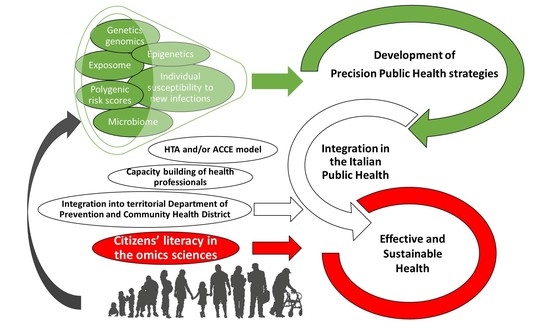
:1. Introduction
2. Development and Perspectives for Italian Public Health Genomic and Epigenomic Tools
2.1. Promising Perspectives from Clinical Genetics: The Use of Polygenic Scores and Epigenetic Markers
2.1.1. Genetics
2.1.2. Epigenetics
2.2. Susceptibility to Environmental Pollution, as Inferred from a miRNA Analysis
2.3. Nutritional and Molecular Epidemiology for Precision Prevention and Health Promotion
2.4. The Microbiome of Children: Development and Disease Implications, and Challenges for a Healthy Life
2.5. A Precision Medicine Approach in COVID-19 Patients: Which Markers Should Be Used for Prognosis?
2.6. Health Technology Assessment for Public Health Evaluation of Genetic/Genomic Applications on Genetic Tests
2.7. Fostering the Implementation of Personalized Healthcare by Developing Health Professionals’ and Citizens’ Omics Science Literacy
- developing awareness among stakeholders;
- improving citizens’ health literacy to fully empower them;
- fostering health professionals’ skills acquisition through extensive educational initiatives in omics sciences;
- shaping sustainable healthcare through the use of evidence-based tools such as a Health Technology Assessment for the omics technologies’ evaluation to introduce in healthcare systems.
2.8. The Point of View of the Territorial Department of Prevention and the Community Health District
- are addressed to healthy people in large numbers;
- represent “proactive” medicine;
- provide cost-effective and evidence-based technologies;
- deliver free or co-payment health care services;
- consider individual as well as community health gain; and
- are provided in all regions of Italy, as they are mandatory.
- cancer screenings
- vaccination campaigns
- risk communication, counseling, health literacy, and empowerment of the target population
- epidemiological evaluation of the health efficacy in the target population
- health surveillance activity
- infectious disease (nowadays, especially COVID-19)
- must be efficacy- and value-based;
- needs dedicated resources (personnel, places, technology, software, etc.);
- needs a structured plan from prevention to treatment;
- must avoid inequalities; and
- requires people’s advocacy and involvement.
- generating more specific and cost-effective public prevention programs;
- enhancing the impact of prevention and risk-reduction campaigns;
- favoring the exchange of information between various branches of the public health sector; and
- maintaining the importance of a central public health author even if the trend is toward personalized medicine.
- consolidated and experienced activity of screening;
- an existing network with clinical disciplines;
- experience of risk communication and counseling;
- experience of follow-up management;
- an appropriate attitude toward the analysis and evaluation of prevention activities;
- appropriate software in use.
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gwinn, M.; MacCannell, D.R.; Khabbaz, R.F. Integrating Advanced Molecular Technologies into Public Health. J. Clin. Microbiol. 2017, 55, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Williamson, D.A.; Kirk, M.D.; Sintchenko, V.; Howden, B.P. The importance of public health genomics for ensuring health security for Australia. Med. J. Aust. 2019, 210, 295–297.e1. [Google Scholar] [CrossRef] [PubMed]
- Ladd-Acosta, C.; Fallin, M.D. The role of epigenetics in genetic and environmental epidemiology. Epigenomics 2016, 8, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Kutateladze, T.G. Diet and the epigenome. Nat. Commun. 2018, 9, 9–11. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Fiore, V.; Rosta, G.; Favara, G.; La Mastra, C.; La Rosa, M.C.; San Lio, R.M.; Agodi, A. Determinants of adherence to the mediterranean diet: Findings from a cross-sectional study in women from Southern Italy. Int. J. Env. Res. Public Health 2019, 16, 2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maugeri, A.; Barchitta, M.; Agrifoglio, O.; Favara, G.; La Mastra, C.; La Rosa, M.C.; Lio, R.M.S.; Panella, M.; Cianci, A.; Agodi, A. The impact of social determinants and lifestyles on dietary patterns during pregnancy: Evidence from the “Mamma & Bambino” study. Ann. Di Ig. 2019, 31, 81–89. [Google Scholar]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Shrestha, G.S.; Paneru, H.R.; Vincent, J.L. Precision medicine for COVID-19: A call for better clinical trials. Crit. Care 2020, 24, 1–3. [Google Scholar] [CrossRef]
- Alberg, C.; Burton, H.; Hall, A.; Wright, C.; Zimmern, R. An Independent Response to the House of Lords Science and Technology Committee Report. PHG Found. 2010, May, 12–14. [Google Scholar]
- Boccia, S.; Federici, A.; Colotto, M.; Villari, P. Implementation of Italian guidelines on public health genomics in Italy: A challenging policy of the NHS TT-Le policy di genomica in sanità pubblica in Italia: Le sfide nella implementazione delle linee guida nel sistema sanitario nazionale. Epidemiol. Prev. 2014, 38, 29–34. [Google Scholar]
- Burke, W.; Atkins, D.; Gwinn, M.; Guttmacher, A.; Haddow, J.; Lau, J.; Palomaki, G.; Press, N.; Richards, C.S.; Wideroff, L.; et al. Genetic test evaluation: Information needs of clinicians, policy makers, and the public. Am. J. Epidemiol. 2002, 156, 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragam, K.G.; Natarajan, P. Polygenic Scores to Assess Atherosclerotic Cardiovascular Disease Risk: Clinical Perspectives and Basic Implications. Circ. Res. 2020, 126, 1159–1177. [Google Scholar] [CrossRef]
- Hachiya, T.; Hata, J.; Hirakawa, Y.; Yoshida, D.; Furuta, Y.; Kitazono, T.; Shimizu, A.; Ninomiya, T. Genome-wide polygenic score and the risk of ischemic stroke in a prospective cohort: The Hisayama study. Stroke 2020, 51, 759–765. [Google Scholar] [CrossRef]
- Ho, W.K.; Tan, M.M.; Mavaddat, N.; Tai, M.C.; Mariapun, S.; Li, J.; Ho, P.J.; Dennis, J.; Tyrer, J.P.; Bolla, M.K.; et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Padilla-Martínez, F.; Collin, F.; Kwasniewski, M.; Kretowski, A. Systematic review of polygenic risk scores for type 1 and type 2 diabetes. Int. J. Mol. Sci. 2020, 21, 1703. [Google Scholar] [CrossRef] [Green Version]
- Ibanez, L.; Farias, F.H.G.; Dube, U.; Mihindukulasuriya, K.A.; Harari, O. Polygenic Risk Scores in Neurodegenerative Diseases: A Review. Curr. Genet. Med. Rep. 2019, 7, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Wendt, F.R.; Pathak, G.A.; Tylee, D.S.; Goswami, A.; Polimanti, R. Heterogeneity and Polygenicity in Psychiatric Disorders: A Genome-Wide Perspective. Chronic Stress 2020, 4. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Gil, L.; Jupp, S.; Ritchie, S.; Xu, Y.; Buniello, A.; Abraham, G.; Chapman, M.; Parkinson, H.; Danesh, J.; et al. The Polygenic Score Catalog: An open database for reproducibility and systematic evaluation. medRxiv 2020. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Kanai, M.; Karjalainen, J.; Akiyama, M.; Kurki, M.; Matoba, N.; Takahashi, A.; Hirata, M.; Kubo, M.; Matsuda, K.; et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat. Med. 2020, 26, 542–548. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M. Meta-analysis of genome-wide association studies for height and body mass index in ~700 000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; Van Der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef] [Green Version]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.R.; Farh, K.H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H.; et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar]
- Gialluisi, A.; Andlauer, T.F.M.; Mirza-Schreiber, N.; Moll, K.; Becker, J.; Hoffmann, P.; Ludwig, K.U.; Czamara, D.; Pourcain, B.S.; Honbolygó, F.; et al. Genome-wide association study reveals new insights into the heritability and genetic correlates of developmental dyslexia. Mol. Psychiatry 2020. [Google Scholar] [CrossRef] [PubMed]
- Yanes, T.; McInerney-Leo, A.M.; Law, M.H.; Cummings, S. The emerging field of polygenic risk scores and perspective for use in clinical care. Hum. Mol. Genet. 2020, 29, R165–R176. [Google Scholar] [CrossRef] [PubMed]
- Hynninen, Y.; Linna, M.; Vilkkumaa, E. Value of genetic testing in the prevention of cardiovascular events (unpublished). PLoS ONE 2019, 14, e0210010. [Google Scholar] [CrossRef] [Green Version]
- Arnett, D.K.; Blumenthal, R.S.; Albert, M.A.; Buroker, A.B.; Goldberger, Z.D.; Hahn, E.J.; Himmelfarb, C.D.; Khera, A.; Lloyd-Jones, D.; McEvoy, J.W.; et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019, 74, e177–e232. [Google Scholar] [CrossRef]
- Piepoli, M.F.; Hoes, A.W.; Agewall, S.; Albus, C.; Brotons, C.; Catapano, A.L.; Cooney, M.T.; Corrà, U.; Cosyns, B.; Deaton, C.; et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice. Atherosclerosis 2016, 252, 207–274. [Google Scholar] [CrossRef] [Green Version]
- Elliott, J.; Bodinier, B.; Bond, T.A.; Chadeau-Hyam, M.; Evangelou, E.; Moons, K.G.M.; Dehghan, A.; Muller, D.C.; Elliott, P.; Tzoulaki, I. Predictive Accuracy of a Polygenic Risk Score-Enhanced Prediction Model vs. a Clinical Risk Score for Coronary Artery Disease. JAMA J. Am. Med. Assoc. 2020, 323, 636–645. [Google Scholar] [CrossRef]
- Läll, K.; Mägi, R.; Morris, A.; Metspalu, A.; Fischer, K. Personalized risk prediction for type 2 diabetes: The potential of genetic risk scores. Genet. Med. 2017, 19, 322–329. [Google Scholar] [CrossRef] [Green Version]
- Pencina, M.J.; D’Agostino, R.B.S.; D’Agostino, R.B.J.; Vasar, R.S. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat. Med. 2008, 27, 157–172. [Google Scholar] [CrossRef]
- Garcìa-Gimènez, J.L.; Seco-Cervera, M.; Tollefsbol, T.O.; Romà-Mateo, C.; Peirò-Chova, L.; Lapuzina, P. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2017, 54, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Relton, C.L.; Hartwig, F.P.; Smith, G.D. From stem cells to the law courts: DNA methylation, the forensic epigenome and the possibility of a biosocial archive. Int. J. Epidemiol. 2015, 44, 1083–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, A.M.; Dogan, M.V.; Beach, S.R.H.; Philibert, R.A. Current and future prospects for epigenetic biomarkers of substance use disorders. Genes 2015, 6, 991–1022. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Mena-Mollá, S.; Beltrán-García, J.; Sanchis-Gomar, F. Challenges in the analysis of epigenetic biomarkers in clinical samples. Clin. Chem. Lab. Med. 2017, 55, 1474–1477. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kaèaèb, S.; Wakili, R.; et al. Stability of circulating blood-based microRNAs-Pre-Analytic methodological considerations. PLoS ONE 2017, 12, 1–16. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [Green Version]
- Zubakov, D.; Boersma, A.W.M.; Choi, Y.; Van Kuijk, P.F.; Wiemer, E.A.C.; Kayser, M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Leg. Med. 2010, 124, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiró-Chova, L.; Peña-Chilet, M.; López-Guerrero, J.A.; García-Giménez, J.L.; Alonso-Yuste, E.; Burgues, O.; Lluch, A.; Ferrer-Lozano, J.; Ribas, G. High stability of microRNAs in tissue samples of compromised quality. Virchows Arch. 2013, 463, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.K.; Mallick, R.; Yendamuri, S. Detection of microRNAs in dried serum blots. Anal. Biochem. 2010, 407, 147–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Giménez, J.L.; Ushijima, T.; Tollefsbol, T.O. Epigenetic Biomarkers: New Findings, Perspectives, and Future Directions in Diagnostics. In Epigenetic Biomarkers and Diagnostics; Elvesier Inc., Ed.; Elvesier: Amsterdam, The Netherlands, 2016; pp. 1–18. [Google Scholar]
- Berdasco, M.; Esteller, M. Aberrant Epigenetic Landscape in Cancer: How Cellular Identity Goes Awry. Dev. Cell 2010, 19, 698–711. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.N.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Bormann, F.; Rodríguez-Paredes, M.; Lasitschka, F.; Edelmann, D.; Musch, T.; Benner, A.; Bergman, Y.; Dieter, S.M.; Ball, C.R.; Glimm, H.; et al. Cell-of-Origin DNA Methylation Signatures Are Maintained during Colorectal Carcinogenesis. Cell Rep. 2018, 23, 3407–3418. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.W.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Paredes, M.; Bormann, F.; Raddatz, G.; Gutekunst, J.; Lucena-Porcel, C.; Köhler, F.; Wurzer, E.; Schmidt, K.; Gallinat, S.; Wenck, H.; et al. Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin. Nat. Commun. 2018, 9, 577. [Google Scholar] [CrossRef]
- Lamb, Y.N.; Dhillon, S. Epi proColon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol. Diagn. 2017, 21, 225–232. [Google Scholar] [CrossRef]
- Ned, R.M.; Melillo, S.; Marrone, M. Fecal DNA testing for Colorectal Cancer Screening: The ColoSureTM test. PLoS Curr. 2011, 22, RRN1220. [Google Scholar] [CrossRef]
- Qazi, T.J.; Quan, Z.; Mir, A.; Qing, H. Epigenetics in Alzheimer’s Disease: Perspective of DNA Methylation. Mol. Neurobiol. 2018, 55, 1026–1044. [Google Scholar] [CrossRef]
- Jakubowski, J.L.; Labrie, V. Epigenetic Biomarkers for Parkinson’s Disease: From Diagnostics to Therapeutics. J. Parkinson’s Dis. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paez-Colasante, X.; Figueroa-Romero, C.; Sakowski, S.A.; Goutman, S.A.; Feldman, E.L. Amyotrophic lateral sclerosis: Mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol. 2015, 11, 266–279. [Google Scholar] [CrossRef]
- Khavari, B.; Cairns, M.J. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells 2020, 9, 1837. [Google Scholar] [CrossRef] [PubMed]
- Penner-Goeke, S.; Binder, E.B. Epigenetics and depression. Dialogues Clin. Neurosci. 2019, 21, 397–405. [Google Scholar] [PubMed]
- Howie, H.; Rijal, C.M.; Ressler, K.J. A review of epigenetic contributions to post-traumatic stress disorder. Dialogues Clin. Neurosci. 2019, 21, 417–428. [Google Scholar] [PubMed]
- Nilsson, E.; Jansson, P.A.; Perfilyev, A.; Volkov, P.; Pedersen, M.; Svensson, M.K.; Poulsen, P.; Ribel-Madsen, R.; Pedersen, N.L.; Almgren, P.; et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 2014, 63, 2962–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkmar, M.; Dedeurwaerder, S.; Cunha, D.A.; Ndlovu, M.N.; Defrance, M.; Deplus, R.; Calonne, E.; Volkmar, U.; Igoillo-Esteve, M.; Naamane, N.; et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012, 31, 1405–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitert, M.D.; Dayeh, T.; Volkov, P.; Elgzyri, T.; Hall, E.; Nilsson, E.; Yang, B.T.; Lang, S.; Parikh, H.; Wessman, Y.; et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 2012, 61, 3322–3332. [Google Scholar] [CrossRef] [Green Version]
- Impact of Overfeeding and Following Exercise Training in Individuals with and without Increased Risk of Type 2 Diabetes-Full Text View-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02982408 (accessed on 6 December 2020).
- Exercise Resistance in Type 2 Diabetes-Full Text View-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT01911104 (accessed on 6 December 2020).
- Talens, R.P.; Jukema, J.W.; Trompet, S.; Kremer, D.; Westendorp, R.G.J.; Lumey, L.H.; Sattar, N.; Putter, H.; Slagboom, P.E.; Heijmans, B.T. Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int. J. Epidemiol. 2012, 41, 106–115. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Ozanne, S.E. Nutrition in early life and age-associated diseases. Ageing Res. Rev. 2017, 39, 96–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freson, K.; Izzi, B.; Labarque, V.; Van Helvoirt, M.; Thys, C.; Wittevrongel, C.; Bex, M.; Bouillon, R.; Godefroid, N.; Proesmans, W.; et al. GNAS defects identified by stimulatory G protein α-subunit signalling studies in platelets. J. Clin. Endocrinol. Metab. 2008, 93, 4851–4859. [Google Scholar] [CrossRef] [Green Version]
- Izzi, B.; Van Geet, C.; Freson, K. Recent Advances in GNAS Epigenetic Research of Pseudohypoparathyroidism. Curr. Mol. Med. 2012, 12, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Freson, K.; Izzi, B.; Van Geet, C. From genetics to epigenetics in platelet research. Thromb. Res. 2012, 129, 325–329. [Google Scholar] [CrossRef]
- Noro, F.; Gianfagna, F.; Gialluisi, A.; De Curtis, A.; Di Castelnuovo, A.; Napoleone, E.; Cerletti, C.; Donati, M.B.; De Gaetano, G.; Hoylaerts, M.F.; et al. ZBTB12 DNA methylation is associated with coagulation- and inflammation-related blood cell parameters: Findings from the Moli-family cohort. Clin. Epigenetics 2019, 11, 1–10. [Google Scholar] [CrossRef]
- Guarrera, S.; Fiorito, G.; Onland-Moret, N.C.; Russo, A.; Agnoli, C.; Allione, A.; Di Gaetano, C.; Mattiello, A.; Ricceri, F.; Chiodini, P.; et al. Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clin. Epigenetics 2015, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Istas, G.; Declerck, K.; Pudenz, M.; Szic, K.S.V.; Lendinez-Tortajada, V.; Leon-Latre, M.; Heyninck, K.; Haegeman, G.; Casasnovas, J.A.; Tellez-Plaza, M.; et al. Identification of differentially methylated BRCA1 and CRISP2 DNA regions as blood surrogate markers for cardiovascular disease. Sci. Rep. 2017, 7, 1–14. [Google Scholar]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Izzotti, A.; Carozzo, S.; Pulliero, A.; Zhabayeva, D.; Ravetti, J.L.; Bersimbaev, R. Extracellular MicroRNA in liquid biopsy: Applicability in cancer diagnosis and prevention. Am. J. Cancer Res. 2016, 6, 1461–1493. [Google Scholar]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Li, Q.; Chen, Q.; Zhou, Y.; Park, S.; Lee, G.; Grimes, B.; Krysan, K.; Yu, M.; Wang, W.; et al. CancerLocator: Non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 2017, 18, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zemmour, H.; Planer, D.; Magenheim, J.; Moss, J.; Neiman, D.; Gilon, D.; Korach, A.; Glaser, B.; Shemer, R.; Landesberg, G.; et al. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat. Commun. 2018, 9, 1443. [Google Scholar] [CrossRef]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef] [PubMed]
- Marotta, A.; Noro, F.; Parisi, R.; Gialluisi, A.; Tirozzi, A.; De Curtis, A.; Costanzo, S.; Di Castelnuovo, A.; Cerletti, C.; Donati, M.B.; et al. NMU DNA methylation in blood is associated with metabolic and inflammatory indices: Results from the Moli-sani study. Epigenetics 2021, 17, 1–14. [Google Scholar] [CrossRef]
- Cohen, A.J.; Brauer, M.; Burnett, R.; Anderson, H.R.; Frostad, J.; Estep, K.; Balakrishnan, K.; Brunekreef, B.; Dandona, L.; Dandona, R.; et al. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: An analysis of data from the Global Burden of Diseases Study 2015. Lancet 2017, 389, 1907–1918. [Google Scholar] [CrossRef] [Green Version]
- Kotsyfakis, M.; Patelarou, E. MicroRNAs as biomarkers of harmful environmental and occupational exposures: A systematic review. Biomarkers 2019, 24, 623–630. [Google Scholar] [CrossRef]
- Izzotti, A.; Pulliero, A. The effects of environmental chemical carcinogens on the microRNA machinery. Int. J. Hyg. Env. Health 2014, 217, 601–627. [Google Scholar] [CrossRef]
- Cheng, M.; Wang, B.; Yang, M.; Ma, J.; Ye, Z.; Xie, L.; Zhou, M.; Chen, W. microRNAs expression in relation to particulate matter exposure: A systematic review. Env. Pollut. 2020, 260, 113961. [Google Scholar] [CrossRef]
- Burris, H.H.; Baccarelli, A.A. Environmental epigenetics: From novelty to scientific discipline. J. Appl. Toxicol. 2014, 34, 113–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duforestel, M.; Briand, J.; Bougras-Carton, G.; Heymann, D.; Frenel, J.-S.; Vallette, F.M.; Carton, P.-F. Cell-free circulating epimarks in cancer monitoring. Epigenomics 2020, 12, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Calin, G.A.; Arrigo, P.; Steele, V.E.; Croce, C.M.; De Flora, S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. Faseb J. 2009, 23, 806–812. [Google Scholar] [CrossRef]
- Schembri, F.; Sridhar, S.; Perdomo, C.; Gustafson, A.M.; Zhang, X.; Ergun, A.; Lu, J.; Liu, G.; Zhang, X.; Bowers, J.; et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 2319–2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izzotti, A.; Larghero, P.; Longobardi, M.; Cartiglia, C.; Camoirano, A.; Steele, V.E.; De Flora, S. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat. Res-Fundam. Mol. Mech. Mutagen. 2011, 717, 9–16. [Google Scholar] [CrossRef]
- Izzotti, A.; Calin, G.A.; Steele, V.E.; Croce, C.M.; De Flora, S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. Faseb J. 2009, 23, 3243–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izzotti, A.; Balansky, R.; Ganchev, G.; Iltcheva, M.; Longobardi, M.; Pulliero, A.; Geretto, M.; Micale, R.T.; La Maestra, S.; Miller, M.S.; et al. Blood and lung microRNAs as biomarkers of pulmonary tumorigenesis in cigarette smoke-exposed mice. Oncotarget 2016, 7, 84758–84774. [Google Scholar] [CrossRef] [Green Version]
- Pulliero, A.; Fazzi, E.; Cartiglia, C.; Orcesi, S.; Balottin, U.; Uggetti, C.; La Piana, R.; Olivieri, I.; Galli, J.; Izzotti, A. The Aicardi-Goutières syndrome. Molecular and clinical features of RNAse deficiency and microRNA overload. Mutat. Res-Fundam. Mol. Mech. Mutagen. 2011, 717, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Pan, Y.; Li, W.; Sun, C.; Liu, J.; Xu, T.; Shu, Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 1–60. [Google Scholar] [CrossRef] [Green Version]
- Kadota, T.; Yoshioka, Y.; Fujita, Y.; Kuwano, K.; Ochiya, T. Extracellular vesicles in lung cancer—From bench to bedside. Semin. Cell Dev. Biol. 2017, 67, 39–47. [Google Scholar] [CrossRef]
- Di Renzo, L.; Gualtieri, P.; Romano, L.; Marrone, G.; Noce, A.; Pujia, A.; Perrone, M.A.; Aiello, V.; Colica, C.; De Lorenzo, A. Role of personalized nutrition in chronic-degenerative diseases. Nutrients 2019, 11, 1707. [Google Scholar] [CrossRef] [Green Version]
- Strianese, O.; Rizzo, F.; Ciccarelli, M.; Galasso, G.; D’agostino, Y.; Salvati, A.; Del Giudice, C.; Tesorio, P.; Rusciano, M.R. Precision and personalized medicine: How genomic approach improves the management of cardiovascular and neurodegenerative disease. Genes 2020, 11, 747. [Google Scholar] [CrossRef]
- Wild, C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [Green Version]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Barone, G.; Mazzoleni, P.; Catalfo, A.; De Guidi, G.; Iemmolo, M.; Crimi, N.; Agodi, A. Integrated approach of nutritional and molecular epidemiology, mineralogical and chemical pollutant characterisation: The protocol of a cross-sectional study in women. BMJ Open 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Agodi, A. The Role of miRNAs as Biomarkers for Pregnancy Outcomes: A Comprehensive Review. Int. J. Genom. 2017, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Vinciguerra, M.; Agodi, A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: A systematic review and meta-analysis. PLoS ONE 2014, 9, e109478. [Google Scholar] [CrossRef]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Canto, C.; La Rosa, N.; Cantarella, M.A.; Spampinato, G.; Scalisi, A.; Agodi, A. LINE-1 hypermethylation in white blood cell DNA is associated with high-grade cervical intraepithelial neoplasia. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Canto, C.; Marchese, A.E.; Vinciguerra, M. Low fruit consumption and folate deficiency are associated with LINE-1 hypomethylation in women of a cancer-free population. Genes Nutr. 2015, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Scalisi, A.; Agodi, A. The association of dietary patterns with high-risk human papillomavirus infection and cervical cancer: A cross-sectional study in Italy. Nutrients 2018, 10, 469. [Google Scholar] [CrossRef] [Green Version]
- Barchitta, M.; Maugeri, A.; La Mastra, C.; La Rosa, M.C.; Favara, G.; Lio, R.M.S.; Agodi, A. Dietary antioxidant intake and human papillomavirus infection: Evidence from a cross-sectional study in Italy. Nutrients 2020, 12, 1384. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Cipresso, R.; Marzagalli, R.; La Rosa, N.; Caruso, M.; Castiglione, M.G.; Travali, S. Distribution of p53, GST, and MTHFR polymorphisms and risk of cervical intraepithelial lesions in sicily. Int. J. Gynecol. Cancer 2010, 20, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Simone, B.; Mazzucco, W.; Gualano, M.R.; Agodi, A.; Coviello, D.; Dagna Bricarelli, F.; Dallapiccola, B.; Di Maria, E.; Federici, A.; Genuardi, M.; et al. The policy of public health genomics in Italy. Health Policy (N.Y.) 2013, 110, 214–219. [Google Scholar] [CrossRef]
- Hibler, E.; Huang, L.; Andrade, J.; Spring, B. Impact of a diet and activity health promotion intervention on regional patterns of DNA methylation. Clin. Epigenetics 2019, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ewald, H.A.S.; Ewald, P.W. Natural selection, microbiomes, and public health. Yale J. Biol. Med. 2018, 91, 1–34. [Google Scholar]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, M.R.; Mitchell, P.D.; Alexander, E.; Ballal, S.; Bartlett, M.; Becker, P.; Davidovics, Z.; Docktor, M.; Dole, M.; Felix, G.; et al. Efficacy of Fecal Microbiota Transplantation for Clostridium difficile Infection in Children. Clin. Gastroenterol. Hepatol. 2020, 18, 612–619.e1. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, J.; Xu, J.; Wei, X.; Yang, J.; Liu, Y.; Li, H.; Zhao, C.; Wang, Y.; Zhang, L.; et al. Helicobacter pylori Infection Aggravates Dysbiosis of Gut Microbiome in Children with Gastritis. Front. Cell. Infect. Microbiol. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhu, Y.; Liao, Q.; Wang, Z.; Wan, C. Characterization of gut microbiota in children with pulmonary tuberculosis. BMC Pediatr. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Nirmalkar, K.; Murugesan, S.; Pizano-Zárate, M.L.; Villalobos-Flores, L.E.; García-González, C.; Morales-Hernández, R.M.; Nuñez-Hernández, J.A.; Hernández-Quiroz, F.; Romero-Figueroa, M.D.S.; Hernández-Guerrero, C.; et al. Gut microbiota and endothelial dysfunction markers in obese Mexican children and adolescents. Nutrients 2018, 10, 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leiva-Gea, I.; Sánchez-Alcoholado, L.; Martín-Tejedor, B.; Castellano-Castillo, D.; Moreno-Indias, I.; Urda-Cardona, A.; Tinahones, F.J.; Fernández-García, J.C.; Queipo-Ortuño, M.I. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: A case-control study. Diabetes Care 2018, 41, 2385–2395. [Google Scholar] [CrossRef] [Green Version]
- Traversi, D.; Rabbone, I.; Scaioli, G.; Vallini, C.; Carletto, G.; Racca, I.; Ala, U.; Durazzo, M.; Collo, A.; Ferro, A.; et al. Risk factors for type 1 diabetes, including environmental, behavioural and gut microbial factors: A case–control study. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Schwimmer, J.B.; Johnson, J.S.; Angeles, J.E.; Behling, C.; Belt, P.H.; Borecki, I.; Bross, C.; Durelle, J.; Goyal, N.P.; Hamilton, G.; et al. Microbiome Signatures Associated with Steatohepatitis and Moderate to Severe Fibrosis in Children with Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 157, 1109–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depner, M.; Taft, D.H.; Kirjavainen, P.V.; Kalanetra, K.M.; Karvonen, A.M.; Peschel, S.; Schmausser-Hechfellner, E.; Roduit, C.; Frei, R.; Lauener, R.; et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat. Med. 2020, 26, 1766–1775. [Google Scholar] [CrossRef]
- Carissimi, C.; Laudadio, I.; Palone, F.; Fulci, V.; Cesi, V.; Cardona, F.; Alfonsi, C.; Cucchiara, S.; Isoldi, S.; Stronati, L. Functional analysis of gut microbiota and immunoinflammation in children with autism spectrum disorders. Dig. Liver Dis. 2019, 51, 1366–1374. [Google Scholar] [CrossRef]
- Sahly, N.; Moustafa, A.; Zaghloul, M.; Salem, T.Z. Effect of radiotherapy on the gut microbiome in pediatric cancer patients: A pilot study. PeerJ 2019, 7, 1–17. [Google Scholar] [CrossRef]
- Li, L.; Wang, F.; Liu, Y.; Gu, F. Intestinal microbiota dysbiosis in children with recurrent respiratory tract infections. Microb. Pathog. 2019, 136, 103709. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Del Chierico, F.; Chiriaco, M.; Foligno, S.; Reddel, S.; Salvatori, G.; Cifaldi, C.; Faraci, S.; Finocchi, A.; Rossi, P.; et al. Gut Mucosal and Fecal Microbiota Profiling Combined to Intestinal Immune System in Neonates Affected by Intestinal Ischemic Injuries. Front. Cell. Infect. Microbiol. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Assfalg, R.; Knoop, J.; Hoffman, K.L.; Pfirrmann, M.; Zapardiel-Gonzalo, J.M.; Hofelich, A.; Eugster, A.; Weigelt, M.; Matzke, C.; Reinhardt, J.; et al. Oral insulin immunotherapy in children at risk for type 1 diabetes in a randomized trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Qian, X.; Liu, Y.X.; Ye, X.; Zheng, W.; Lv, S.; Mo, M.; Lin, J.; Wang, W.; Wang, W.; Zhang, X.; et al. Gut microbiota in children with juvenile idiopathic arthritis: Characteristics, biomarker identification, and usefulness in clinical prediction. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Valentini, F.; Evangelisti, M.; Arpinelli, M.; Di Nardo, G.; Borro, M.; Simmaco, M.; Villa, M.P. Gut microbiota composition in children with Obstructive Sleep Apnoea Syndrome: A pilot study. Sleep Med. 2020, 76, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Cen, S.; Jiang, J.; Zhao, J.; Zhang, H.; Chen, W. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: A pilot study of Chinese children. Env. Res. 2019, 171, 501–509. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, J.; He, F.; Cai, C.; Wang, H.; Wang, Y.; Lin, Y.; Rong, H.; Cheng, G.; Xu, R.; et al. Alteration of gut microbiota-associated epitopes in children with autism spectrum disorders. Brain Behav. Immun. 2019, 75, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Yooseph, S.; Harkins, D.M.; Moncera, K.J.; Zabokrtsky, K.B.; Torralba, M.G.; Tovchigrechko, A.; Highlander, S.; Pieper, R.; Sender, L.; et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genom. 2016, 17, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, D.J.; Reynolds, R.M.; Hardy, D.B. Developmental origins of health and disease: Current knowledge and potential mechanisms. Nutr. Rev. 2017, 75, 951–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilonen, J.; Lempainen, J.; Veijola, R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 635–650. [Google Scholar] [CrossRef]
- Wang, S.; Xu, M.; Wang, W.; Cao, X.; Piao, M.; Khan, S.; Yan, F.; Cao, H.; Wang, B. Systematic review: Adverse events of fecal Microbiota transplantation. PLoS ONE 2016, 11, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef]
- Javdan, B.; Lopez, J.G.; Chankhamjon, P.; Lee, Y.C.J.; Hull, R.; Wu, Q.; Wang, X.; Chatterjee, S.; Donia, M.S. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 2020, 181, 1661–1679.e22. [Google Scholar] [CrossRef] [PubMed]
- Sagner, M.; McNeil, A.; Puska, P.; Auffray, C.; Price, N.D.; Hood, L.; Lavie, C.J.; Han, Z.G.; Chen, Z.; Brahmachari, S.K.; et al. The P4 Health Spectrum–A Predictive, Preventive, Personalized and Participatory Continuum for Promoting Healthspan. Prog. Cardiovasc. Dis. 2017, 59, 506–521. [Google Scholar] [CrossRef] [Green Version]
- Crisci, C.D.; Ardusso, L.R.F.; Mossuz, A.; Müller, L. A Precision Medicine Approach to SARS-CoV-2 Pandemic Management. Curr. Treat. Options Allergy 2020, 7, 422–440. [Google Scholar] [CrossRef]
- Sigman, M. Introduction: Personalized medicine: What is it and what are the challenges? Fertil. Steril. 2018, 109, 944–945. [Google Scholar] [CrossRef]
- Prokop, J.W.; Shankar, R.; Gupta, R.; Leimanis, M.L.; Nedveck, D.; Uhl, K.; Chen, B.; Hartog, N.L.; Van Veen, J.; Sisco, J.S.; et al. Virus-induced genetics revealed by multidimensional precision medicine transcriptional workflow applicable to COVID-19. Physiol. Genom. 2020, 52, 255–268. [Google Scholar] [CrossRef]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B.; Haralambieva, I.H.; Jacobson, R.M. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. Omics. 2011, 15, 625–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamburg, M.A.; Collins, F.S. The path to personalized medicine. N. Engl. J. Med. 2010, 363, 301–304. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, I.V. The collective nature of personalized medicine. Genet. Res. (Camb) 2016, 98, e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shomron, N. Prioritizing personalized medicine. Genet. Res. (Camb) 2014, 96, 19–20. [Google Scholar] [CrossRef] [Green Version]
- Poland, G.A. Pharmacology, vaccinomics, and the second golden age of vaccinology. Clin. Pharm. 2007, 82, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. Personalized vaccinology: A review. Vaccine 2018, 36, 5350–5357. [Google Scholar] [CrossRef]
- Wollenstein-Betech, S.; Cassandras, C.G.; Paschalidis, I.C. Personalized predictive models for symptomatic COVID-19 patients using basic preconditions: Hospitalizations, mortality, and the need for and ICU or ventilator. Int. J. Med. Inform. 2020, 123, 11–22. [Google Scholar] [CrossRef]
- Baj, J.; Karakuła-Juchnowicz, H.; Teresiński, G.; Buszewicz, G.; Ciesielka, M.; Sitarz, E.; Forma, A.; Karakuła, K.; Flieger, W.; Portincasa, P.; et al. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J. Clin. Med. 2020, 9, 1753. [Google Scholar] [CrossRef]
- Rubino, S.; Kelvin, N.; Bermejo-Martin, J.F.; Kelvin, D.J. As COVID-19 cases, deaths and fatality rates surge in Italy, underlying causes require investigation. J. Infect. Dev. Ctries. 2020, 14, 265–267. [Google Scholar] [CrossRef] [Green Version]
- Cesari, M.; Montero-Odasso, M. COVID-19 and older adults. lessons learned from the Italian epicenter. Can. Geriatr. J. 2020, 23, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Iosa, M.; Paolucci, S.; Morone, G. Covid-19: A Dynamic Analysis of Fatality Risk in Italy. Front. Med. 2020, 7, 1–5. [Google Scholar] [CrossRef]
- Onder, G.; Rezza, G.; Brusaferro, S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. Jama J. Am. Med. Assoc. 2020, 323, 1775–1776. [Google Scholar] [CrossRef] [PubMed]
- Parisi, V.; Leosco, D. Precision Medicine in COVID-19: IL-1β a Potential Target. Jacc Basic Transl. Sci. 2020, 5, 543–544. [Google Scholar] [CrossRef] [PubMed]
- Sepulchre, E.; Pittie, G.; Stojkovic, V.; Haesbroek, G.; Crama, Y.; Schyns, M.; Paridaens, H.; de Marchin, J.; Degesves, S.; Biemar, C.; et al. Covid-19: Contribution of clinical characteristics and laboratory features for early detection of patients with high risk of severe evolution. Acta Clin. Belg. Int. J. Clin. Lab. Med. 2020, 1–7. [Google Scholar] [CrossRef]
- Ray, S.; Srivastava, S. COVID-19 pandemic: Hopes from proteomics and multiomics research. Omics. 2020, 24, 457–459. [Google Scholar] [CrossRef]
- Ahmed, Z. Practicing precision medicine with intelligently integrative clinical and multi-omics data analysis. Hum. Genom. 2020, 14, 1–5. [Google Scholar] [CrossRef]
- Eckhardt, M.; Hultquist, J.F.; Kaake, R.M.; Hüttenhain, R.; Krogan, N.J. A systems approach to infectious disease. Nat. Rev. Genet. 2020, 21, 339–354. [Google Scholar] [CrossRef]
- Omersel, J.; Karas Kuželički, N. Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines. J. Clin. Med. 2020, 9, 3561. [Google Scholar] [CrossRef] [PubMed]
- Bhopal, S.S.; Bhopal, R. Sex differential in COVID-19 mortality varies markedly by age. Lancet 2020, 396, 532–533. [Google Scholar] [CrossRef]
- Michelozzi, P.; De’Donato, F.; Scortichini, M.; De Sario, M.; Noccioli, F.; Rossi, P.; Davoli, M. Mortality impacts of the coronavirus disease (COVID-19) outbreak by sex and age: Rapid mortality surveillance system, Italy, 1 February to 18 April 2020. Eurosurveillance 2020, 25, 1–5. [Google Scholar] [CrossRef]
- Franchini, M.; Glingani, C.; Del Fante, C.; Capuzzo, M.; Di Stasi, V.; Rastrelli, G.; Vignozzi, L.; De Donno, G.; Perotti, C. The protective effect of O blood type against SARS-CoV-2 infection. Vox Sang. 2020, 1–2. [Google Scholar] [CrossRef]
- Golinelli, D.; Boetto, E.; Maietti, E.; Fantini, M.P. The association between ABO blood group and SARS-CoV-2 infection: A meta-analysis. PLoS ONE 2020, 15, 1–14. [Google Scholar] [CrossRef]
- Sherwani, S.; Khan, M.W.A. Cytokine response in SARS-CoV-2 infection in the Elderly. J. Inflamm. Res. 2020, 13, 737–747. [Google Scholar] [CrossRef]
- Fadini, G.P.; Morieri, M.L.; Boscari, F.; Fioretto, P.; Maran, A.; Busetto, L.; Bonora, B.M.; Selmin, E.; Arcidiacono, G.; Pinelli, S.; et al. Newly-diagnosed diabetes and admission hyperglycemia predict COVID-19 severity by aggravating respiratory deterioration. Diabetes Res. Clin. Pr. 2020, 168, 108374. [Google Scholar] [CrossRef]
- Figliozzi, S.; Masci, P.G.; Ahmadi, N.; Tondi, L.; Koutli, E.; Aimo, A.; Stamatelopoulos, K.; Dimopoulos, M.A.; Caforio, A.L.P.; Georgiopoulos, G. Predictors of adverse prognosis in COVID-19: A systematic review and meta-analysis. Eur. J. Clin. Invest. 2020, 50, 1–15. [Google Scholar] [CrossRef] [PubMed]
- La Vignera, S.; Cannarella, R.; Condorelli, R.A.; Torre, F.; Aversa, A.; Calogero, A.E. Sex-specific SARS-CoV2 mortality: Among hormone-modulated ACE2 expression, risk of venous thromboembolism and hypovitaminosis D. Int. J. Mol. Sci. 2020, 21, 2948. [Google Scholar] [CrossRef]
- Maddaloni, E.; D’Onofrio, L.; Alessandri, F.; Mignogna, C.; Leto, G.; Pascarella, G.; Mezzaroma, I.; Lichtner, M.; Pozzilli, P.; Agrò, F.E.; et al. Cardiometabolic multimorbidity is associated with a worse Covid-19 prognosis than individual cardiometabolic risk factors: A multicentre retrospective study (CoViDiab II). Cardiovasc. Diabetol. 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Ciceri, F.; Castagna, A.; Rovere-Querini, P.; De Cobelli, F.; Ruggeri, A.; Galli, L.; Conte, C.; De Lorenzo, R.; Poli, A.; Ambrosio, A.; et al. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clin. Immunol. 2020, 217, 108509. [Google Scholar] [CrossRef] [PubMed]
- Sieńko, J.; Kotowski, M.; Bogacz, A.; Lechowicz, K.; Drożdżal, S.; Rosik, J.; Sietnicki, M.; Sieńko, M.; Kotfis, K. COVID-19: The influence of ACE genotype and ACE-I and ARBs on the course of SARS-CoV-2 infection in elderly patients. Clin. Interv. Aging 2020, 15, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Ariumi, Y.; Nishida, N.; Yamamoto, R.; Bauer, G.; Gojobori, T.; Shimotohno, K.; Mizokami, M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene 2020, 758, 144944. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- The Sever Covid-19 GWAS Group Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [CrossRef]
- Barnkob, M.B.; Pottegård, A.; Støvring, H.; Haunstrup, T.M.; Homburg, K.; Larsen, R.; Hansen, M.B.; Titlestad, K.; Aagaard, B.; Møller, B.K.; et al. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020, 4, 4990–4993. [Google Scholar] [CrossRef] [PubMed]
- Latz, C.A.; DeCarlo, C.; Boitano, L.; Png, C.Y.M.; Patell, R.; Conrad, M.F.; Eagleton, M.; Dua, A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020, 99, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Hoiland, R.L.; Fergusson, N.A.; Mitra, A.R.; Griesdale, D.E.G.; Devine, D.V.; Stukas, S.; Cooper, J.; Thiara, S.; Foster, D.; Chen, L.Y.C.; et al. The association of ABO blood group with indices of disease severity and multiorgan dysfunction in COVID-19. Blood Adv. 2020, 4, 4981–4989. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T. Relationship between the ABO Blood Group and the COVID-19 Susceptibilit. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [Green Version]
- Focosi, D. Anti-A isohaemagglutinin titres and SARS-CoV-2 neutralization: Implications for children and convalescent plasma selection. Br. J. Haematol. 2020, 190, e148–e150. [Google Scholar] [CrossRef] [PubMed]
- Cooling, L. Blood groups in infection and host susceptibility. Clin. Microbiol. Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020, 12, 1–18. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Pitini, E.; De Vito, C.; Marzuillo, C.; D’Andrea, E.; Rosso, A.; Federici, A.; Di Maria, E.; Villari, P. How is genetic testing evaluated? A systematic review of the literature. Eur. J. Hum. Genet. 2018, 26, 605–615. [Google Scholar] [CrossRef] [Green Version]
- ACCE Model Process for Evaluating Genetic Tests | CDC. Available online: https://www.cdc.gov/genomics/gtesting/acce/index.htm (accessed on 6 December 2020).
- Haddow, J.E.; Palomaki, G. ACCE: A Model Process for Evaluating Data on Emerging Genetic Tests; Khoury, M., Little, J., Burke, W., Eds.; ACCE (CDC USA.gov): Washington, DC, USA, 2004.
- Giacomini, M.; Miller, F.; Browman, G. Confronting the “Gray Zones” of Technology Assessment: Evaluating genetic testing services for public insurance coverage in Canada. Int. J. Technol. Assess. Health Care 2003, 19, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Battista, R.N. Expanding the scientific basis of health technology assessment: A research agenda for the next decade. Int. J. Technol. Assess. Health Care 2006, 22, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Pitini, E.; D’Andrea, E.; De Vito, C.; Rosso, A.; Unim, B.; Marzuillo, C.; Federici, A.; Di Maria, E.; Villari, P. A proposal of a new evaluation framework towards implementation of genetic tests. PLoS ONE 2019, 14, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, J.N.; Turbitt, E.; Biesecker, B.B. Personal utility in genomic testing: A systematic literature review. Eur. J. Hum. Genet. 2017, 25, 662–668. [Google Scholar] [CrossRef]
- Unim, B.; Pitini, E.; Lagerberg, T.; Adamo, G.; De Vito, C.; Marzuillo, C.; Villari, P. Current genetic service delivery models for the provision of genetic testing in Europe: A systematic review of the literature. Front. Genet. 2019, 10, 1–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The EUnetHTA JA 2 HTA Core Model ® Online User guide. Eunethta JA 2015, 2, 1–27.
- NEJM Catalyst What Is Patient-Centered Care? Available online: https://catalyst.nejm.org/doi/full/10.1056/CAT.17.0559 (accessed on 6 December 2020).
- Khoury, M.J. No shortcuts on the long road to evidence-based genomic medicine. Jama J. Am. Med. Assoc. 2017, 318, 27–28. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, W.; Boccia, S. New challenges of public health: Bringing the future of personalised healthcare into focus. Eur. J. Public Health 2017, 27, 36–39. [Google Scholar] [CrossRef]
- Mazzucco, W.; Pastorino, R.; Lagerberg, T.; Colotto, M.; D’Andrea, E.; Marotta, C.; Marzuillo, C.; Villari, P.; Federici, A.; Ricciardi, W.; et al. Current state of genomic policies in healthcare among EU member states: Results of a survey of chief medical officers. Eur. J. Public Health 2017, 27, 931–937. [Google Scholar] [CrossRef] [Green Version]
- Boccia, S.; Federici, A.; Siliquini, R.; Calabrò, G.E.; Ricciardi, W. Implementation of genomic policies in Italy: The new national plan for innovation of the health system based on omics sciences. Epidemiol. Biostat. Public Health 2017. [Google Scholar]
- Boccia, S.; Pastorino, R.; Ricciardi, W.; Ádány, R.; Barnhoorn, F.; Boffetta, P.; Cornel, M.C.; De Vito, C.; Gray, M.; Jani, A.; et al. How to Integrate Personalized Medicine into Prevention? Recommendations from the Personalized Prevention of Chronic Diseases (PRECeDI) Consortium. Public Health Genom. 2019, 22, 208–214. [Google Scholar] [CrossRef]
- Pritchard, D.E.; Moeckel, F.; Villa, M.S.; Housman, L.T.; McCarty, C.A.; McLeod, H.L. Strategies for integrating personalized medicine into healthcare practice. Pers. Med. 2017, 14, 141–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelazzo, M.B.; Pastorino, R.; Mazzucco, W.; Boccia, S. Distance learning training in genetics and genomics testing for Italian health professionals: Results of a pre and post-test evaluation. Epidemiol. Biostat. Public Health 2015, 12, e11516-1–e11516-6. [Google Scholar]
- Calabrò, G.E.; Tognetto, A.; Mazzaccara, A.; Barbina, D.; Carbone, P.; Guerrera, D.; Federici, A.; Ricciardi, W.; Boccia, S. Omic sciences and capacity building of health professionals: A distance learning training course for Italian physicians, 2017–2018. Ig. Sanita Pubblica 2019, 75, 105–124. [Google Scholar]
- Eduiss.it. Available online: https://www.eduiss.it/ (accessed on 12 December 2020).
- Budin-Ljøsne, I.; Harris, J.R. Ask not what personalized medicine can do for you-Ask what you can do for personalized medicine. Public Health Genom. 2015, 18, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Su, P. Direct-to-Consumer Genetic Testing: A Comprehensive View. Yale J. Biol. Med. 2013, 86, 359–365. [Google Scholar]
- Calabrò, G.E.; Sassano, M.; Tognetto, A.; Boccia, S. Citizens’ Attitudes, Knowledge, and Educational Needs in the Field of Omics Sciences: A Systematic Literature Review. Front. Genet. 2020, 11, 570649. [Google Scholar] [CrossRef]
- Boccia, S.; Del Sole, A.; Simone, B.; Sbrogiò, L. Genomica di sanità pubblica e medicina predittiva. Rapp. Prev. 2011, 1, 613–636. [Google Scholar]
- Sbrogiò, L.; Banovich, F.; Favaretto, A.; Vigiani, N.; Del Sole, A. La Medicina Predittiva: Quali Prospettive Operative per i Dipartimenti di Prevenzione; atti 44° Congresso Nazionale S.It.I.: Venezia, Italy, 2010. [Google Scholar]
- Ministero della Salute. Piano Nazionale di Prevenzione 2010–2012; Ministero della Salute: Roma, Italy, 2010.
- Ministero della Salute. Piano Nazionale della Prevenzione 2020–2025; Ministero della Salute: Roma, Italy, 2020.
- Ministero della Salute. Piano nazionale della prevenzione 2014–2018; Ministero della Salute: Roma, Italy, 2015.
- Ministero della Salute. Intesa Stato Regioni del 13 Marzo 2013 “Linee di Indirizzo Sulla Genomica in Sanità Pubblica”; Ministero della Salute: Roma, Italy, 2013.
- Ministero della Salute. Intesa Stato Regioni del 26 Ottobre 2017 “Piano per L’innovazione del Sistema Sanitario Basata Sulle Scienze Omiche”; Ministero della Salute: Roma, Italy, 2017.
- DPCM. Definizione e Aggiornamento dei Livelli Essenziali di Assistenza, di cui All’articolo 1, Comma 7, del Decreto Legislativo 30 Dicembre 1992, n. 502; Ministero della Salute: Roma, Italy, 2017.
| Disease | Age | No. of Participants | Study Design | Results | Biological Plausibility | Author, Year |
|---|---|---|---|---|---|---|
| Clostridium difficile infection (CDI) | 11 months–23 years old | 372 patients (31 were excluded because they had fewer than 60 days of follow-up, six because of refractory CDI) | Cohort study | Fecal Microbiota Transplantation (FMT) had a successful outcome in CDI pediatric patients: 81% had a successful outcome following a single FMT and 86.6% had a successful outcome following a first or repeated FMT; 4.7% had a severe adverse event during the three-month follow-up period, including 10 hospitalizations | There were four independent predictors of FMT success:
| [108] |
| H. pylori-induced gastritis | 4–14 years old | 154 (52 H. pylori-induced gastritis (HPG), 42 H. pylori-negative gastritis (HNG), 62 healthy control (HCG)) | Case-control study | Changes in F:B ratio, an increase of Bacteroidaceae and Enterobacteriaceae, and a decrease of Lachnospiraceae and Bifidobacteriaceae can be caused by gastritis itself and exacerbated by H. pylori infection. These changes may be related to drug resistance and the development of chronic gastrointestinal diseases. | Most of the significant taxa belonged to the Gram-negative bacteria producing LPS. The LPS from the intestinal microbiome induces a chronic subclinical inflammatory process. The upregulation of pro-inflammatory cytokines and downregulation of anti-inflammatory cytokines may be a way to influence gastritis. Lactobacillus can change the pH of the intestinal environment to inhibit the growth of pathogenic bacteria and stimulate an immune response. | [109] |
| Tuberculosis | <14 years old | 36 (18 diagnosed or probably infected + 18 healthy controls) | Case-control study | Pulmonary TB patients presented an upregulation of Prevotella, Enterococcus, and a reduction of Ruminococcaceae, Bifidobacteriaceae, and F. prausnitzii. | Prevotella is a pro-inflammatory bacterium that may activate inflammatory reactions that aggravate TB. Enterococcus is a pathogen associated with intestinal permeability. The translocation of this bacteria into systemic circulation induces an immune-inflammatory reaction. F. prausnitzii is an SFCA producer and SCFAs regulate intestinal permeability. Alterations in Bifidobacteriaceae may be associated with a reduction in the immune response against the invasion of foreign microbes. | [110] |
| Recurrent respiratory tract Infections (RRTI) | Under five years old | 49 (26 patients and 23 healthy controls) | Case-control study | ROC analysis: Enterococcus achieving AUC values of 0.860 | Changes in the gut microbiome’s constituents, with an increased incidence of opportunistic pathogens like Enterococcus, are linked to altered immune responses and homeostasis in the airways. | [118] |
| Intestinal ischemic injuries | <14 years old | 14 patients + 9 healthy controls | Case-control study | Enterobacteriaceae’s and Veillonella dispar’s increase and a reduction in Akkermansia muciniphila might be investigated as a target of intestinal injuries in neonates. | Enterobacteriaceae may be related to a pro-inflammatory response by the immature immune system, resulting in homeostasis disruption. A. muciniphila stimulates in mice the proliferation of Treg cells and is observed in patients with inflammatory bowel disease, suggesting it may have anti-inflammatory properties. Instead, V. dispar has pro-inflammatory effects. | [119] |
| Nonalcoholic Fatty Liver Disease (NAFLD) | 8–17 years old | 124 (87 biopsy-proven NAFLD, 37 obese controls) N.B. NAFLD patients were more likely to be male | Case-control study | Prevotella was more abundant in children with NASH or obesity. | P. copri is the dominant Prevotella species. Data analysis showed that P. copri abundance was the best predictor of fibrosis severity. P. copri increases intestinal inflammation to its advantage. Such pro-inflammatory effects may exacerbate liver damage. | [114] |
| Obesity | 111 children aged 6–11; 61 adolescents aged 12–18 | 172 (76 normal-weight and 96 obese individuals, of whom 46.88% were affected by metabolic syndrome) | Case-control study | Obese children had a higher relative abundance of Firmicutes and Actinobacteria and decreased Bacteroidetes. | Coriobacteriaceae family positively correlates with intrahepatic levels of triglycerides and non-HDL plasma concentrations, suggesting an effect on the gut barrier. Prevotella is associated with chronic inflammation. Firmicutes phylum: Lactobacillus is associated with weight gain. | [111] |
| Type 1 diabetes mellitus (T1D) | Under 18 years old | 15 T1DM + 15 nonautoimmune diabetes + 13 healthy controls | Case-control study | Gut microbiota in T1D differs at the taxonomic and functional levels in comparison with healthy subjects and nonautoimmune diabetes patients. T1D was characterized by an increase in Bacteroidete and pro-inflammatory bacteria, and a decrease in Faecalibacterium and Roseburia. | The T1D gut microbiota profile was associated with a loss of epithelial integrity, low-grade inflammation, and autoimmune response, allowing luminal antigens to escape from the gut and promote islet-directed autoimmune responses. The gut microbiota from patients with T1D was significantly enriched with genes for antigen presentation, chemokine production, LPS biosynthesis, and bacterial invasion. | [112] |
| 2 months‒6 years old | 44 children with a first-degree family history of T1D; 22 were exposed to oral insulin and 22 to a placebo. | Cohort study | There are differences in microbiome composition between children with susceptible and nonsusceptible INS genotypes, and after oral insulin treatment in children with the susceptible INS AA genotype. | There is an increased abundance of Bacteroides dorei in children with the susceptible INS AA genotype. There is an increased alpha diversity in children treated with oral insulin, who showed an antibody response compared with those without a response; this observation is consistent with a microbiome-mediated treatment effect. | [120] | |
| 5–10 years old | 40 T1D patients and 56 healthy children | Case-control study | Modulation of the T1D risk includes higher Firmicutes levels (OR 7.30; IC 2.26–23.54) and a greater amount of Bifidobacterium in the gut (OR 0.13; IC 0.05–0.34) | The origin of the disease process was suspected to be gut microbiota dysbiosis, associated with altered gut permeability and a major vulnerability of the immune system. | [113] | |
| Juvenile idiopathic arthritis (JIA) | 1–16 years old | 39 JIA diagnosed patients + 42 healthy controls | Case-control study | The relative abundance of four genera, Anaerostipes, Dialister, Lachnospira, and Roseburia, decreased significantly in the JIA group. 12 genera were identified as potential biomarkers (AUC = 0.7975): Bifidobacterium, Lachnospira, Dialister, Roseburia, Oscillospira, Akkermansia, Clostridium, Faecalibacterium, Bilophila, Coprococcus, Haemophilus, Anaerostipes. | Anaerostipes, Dialister, Lachnospira, and Roseburia in JIA patients decreased, three of which are butyrate-producing microbes; Dialister is a propionate-producing microbe. SCFAs have considerable immunomodulatory effects (inducing the differentiation of regulatory T cells, enhancing IL-10 production, and suppressing Th17 cells; butyrate administration suppressed the expression of inflammatory cytokines). | [121] |
| Asthma | 2–12 months old | 618 children for bacterial 16S rRNA 189 children for fungal ITS region | Case-control study | There is an inverse association of asthma with the measured level of fecal butyrate (OR = 0.28 (0.09–0.91), P = 0.034), bacterial taxa butyrate producers (Roseburia and Coprococcus, OR = 0.38 (0.17–0.84), P = 0.017) and the relative abundance of the gene encoding butyryl–coenzyme A (CoA): acetate–CoA-transferase, (OR = 0.43 (0.19–0.97), P = 0.042). Children who had grown up on farms had a lower risk of asthma compared to others (OR = 0.56). | Butyrate is the main source of energy for colonic epithelial cells; it contributes to the maintenance of the epithelial gut barrier and has immunomodulatory and anti-inflammatory properties. | [115] |
| Obstructive sleep apnea syndrome (OSAS) | 2–12 years old | 16 (divided between patients and healthy controls) | Case-control study | Faecalibacterium decrease in children with severe grades of OSAS. | Faecalibacterium is involved in the production of butyrate, which improves the gut barrier function, upregulating mucin-associated genes in gut goblet cells and the expression of the tight junction proteins. | [122] |
| Autism spectrum disease (ASD) | 2–6 years old | 16 ASD children + 7 controls | Case-control study | Gut microbiota decreased biodiversity: four of the 82 GO terms have a role in the catabolic process of the 3,3phenylpropionate mapped to the E. coli group. | 3,3phenylpropionate is the conjugate base of 3-phenylpropionic acid deriving from PPA. PPA is an SCFA produced during the bacterial fermentation of carbohydrates. The elevated concentration of propionate metabolites could be due to their reduced degradation because of the E. coli drop. | [116] |
| 3–7 years old | 78 ASD children + 58 controls | Case-control study | Nine genera and the abundance of seven metallic elements are altered in ASD children. These were used in a diagnostic model in Chinese children with high accuracy (84%). | The diagnostic model is composted by bacterial genera (Bacteroides, Parabacteroides, Sutterella, Lachnospira, Bacillus, Bilophila, Lactococcus, Lachnobacterium, and Oscillospira) and metallic elements (Pb, As, Cu, Zn, Mg, Ca, and Hg). Parabacteroides and Oscillospira changes could be induced by heavy metal exposure. | [123] | |
| 2–8 years old | 43 ASD children (19 with GI symptoms and 24 without) + 31 controls | Case-control study | 34 MEs (gut microbiota-associated epitopes) are a potential biomarker of ASD. Those alterations may contribute to abnormalities in gut immunity and/or homeostasis in ASD children. | 29 of 34 MEs decreased and were associated with abnormal gut IgA levels and altered gut microbiota composition;11 of 29 were pathogenic microorganisms’ peptides with T or B cell response. ME with homology to a Listeriolysin O peptide from the pathogenic bacterium Listeria monocytogenes is increased. | [124] | |
| Acute lymphoblastic leukemia (ALL) | 2–25 years old | 51 (23 matched patients and a healthy sibling and five unmatched patients) | Case-control study | It was possible to distinguish between the patient and control groups based on their microbiota profiles. Lachnospiraceae (which comprises the Clostridium XIVa) and Roseburia are butyrate-producing bacteria and were greatly reduced in acute leukemia patients compared to a healthy sibling; instead, Bacteroides increased. | Bacteria producing butyrate play a major role in the composition of the mucus layer, as butyrate is an important energy source for intestinal epithelial cells and plays a role in the maintenance of colonic homeostasis. Butyrate-producing bacteria may increase the risk of developing chemotherapy-induced mucositis and other GI complications. Antibiotic-induced shifts can increase the susceptibility to C. difficile infection. | [125] |
| Rhabdomyosarcoma | 3–7 years old | 3 oncologic patients + 2 healthy controls | Case-control study | After radiation exposure, there was an increase in α-diversity related to nonresponsive radiotherapy treatment, and a decrease in Firmicutes, associated with a Proteobacteria increase. This information could be used for the definition of the therapy. | The decrease of Firmicutes could explain the variation in α-diversity and the ability to survive of the Proteobacteria phylum and might be related to DNA mutations. | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traversi, D.; Pulliero, A.; Izzotti, A.; Franchitti, E.; Iacoviello, L.; Gianfagna, F.; Gialluisi, A.; Izzi, B.; Agodi, A.; Barchitta, M.; et al. Precision Medicine and Public Health: New Challenges for Effective and Sustainable Health. J. Pers. Med. 2021, 11, 135. https://doi.org/10.3390/jpm11020135
Traversi D, Pulliero A, Izzotti A, Franchitti E, Iacoviello L, Gianfagna F, Gialluisi A, Izzi B, Agodi A, Barchitta M, et al. Precision Medicine and Public Health: New Challenges for Effective and Sustainable Health. Journal of Personalized Medicine. 2021; 11(2):135. https://doi.org/10.3390/jpm11020135
Chicago/Turabian StyleTraversi, Deborah, Alessandra Pulliero, Alberto Izzotti, Elena Franchitti, Licia Iacoviello, Francesco Gianfagna, Alessandro Gialluisi, Benedetta Izzi, Antonella Agodi, Martina Barchitta, and et al. 2021. "Precision Medicine and Public Health: New Challenges for Effective and Sustainable Health" Journal of Personalized Medicine 11, no. 2: 135. https://doi.org/10.3390/jpm11020135
APA StyleTraversi, D., Pulliero, A., Izzotti, A., Franchitti, E., Iacoviello, L., Gianfagna, F., Gialluisi, A., Izzi, B., Agodi, A., Barchitta, M., Calabrò, G. E., Hoxhaj, I., Sassano, M., Sbrogiò, L. G., Del Sole, A., Marchiori, F., Pitini, E., Migliara, G., Marzuillo, C., ... Boccia, S. (2021). Precision Medicine and Public Health: New Challenges for Effective and Sustainable Health. Journal of Personalized Medicine, 11(2), 135. https://doi.org/10.3390/jpm11020135