Parathyroid Hormone Gene and Genes Involved in the Maintenance of Vitamin D Levels Association with Mandibular Retrognathism
Abstract
:1. Introduction
2. Materials and Methods
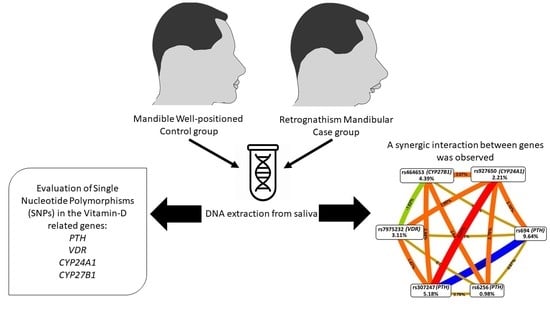
2.1. Sample
2.2. Phenotypes Definition
2.3. Allelic Discrimination
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mossey, P.A. The heritability of malocclusion: Part 2. The influence of genetics in malocclusion. Br. J. Orthod. 1999, 26, 195–203. [Google Scholar] [CrossRef]
- Markovic, M.D. At the crossroads of oral facial genetics. Eur. J. Orthod. 1992, 14, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Lundström, A. Nature versus nurture in dento-facial variation. Eur. J. Orthod. 1994, 6, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Nakasima, A.; Ichinose, M.; Nakata, S.; Takahama, Y. Hereditary factors in the craniofacial morphology of Angle’s Class II and Class III malocclusions. Am. J. Orthod. 1982, 82, 1–26. [Google Scholar] [CrossRef]
- Uribe, L.M.M.; Miller, S.F. Genetics of the dentofacial variation in human malocclusion. Orthod. Craniofac. Res. 2015, 18, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehesa-Santos, A.; Iber-Diaz, P.; Iglesias-Linares, A. Genetic factors contributing to skeletal class III malocclusion: A systematic review and meta-analysis. Clin. Oral Investig. 2021, 25, 1587–1612. [Google Scholar] [CrossRef] [PubMed]
- Balkhande, P.B.; Lakkakula, B.V.K.S.; Chitharanjan, A.B. Relationship between matrilin-1 gene polymorphisms and mandibular retrognathism. Am. J. Orthod. Dentofac. Orthop. 2018, 153, 255–261. [Google Scholar] [CrossRef]
- Teama, S. DNA polymorphisms: DNA-based molecular markers and their application in medicine. In Genetic Diversity and Disease Susceptibility, 1st ed.; Liu, Y., Ed.; IntechOpen: London, UK, 2018; Volume 25. [Google Scholar]
- Arun, R.M.; Lakkakula, B.V.; Chitharanjan, A.B. Role of myosin 1H gene p morphisms in mandibular retrognathism. Am. J. Orthod. Dentofac. Orthop. 2016, 149, 699–704. [Google Scholar] [CrossRef]
- Wang, C.; Ni, Z.; Cai, Y.; Zhou, Y.; Chen, W. Association of Polymorphism rs67920064 in ADAMTS9 Gene with Mandibular Retrognathism in a Chinese Population. Med. Sci. Monit. 2020, 30, e925965. [Google Scholar]
- Küchler, E.C.; Reis, C.L.B.; Carelli, J.; Scariot, R.; Nelson-Filho, P.; Coletta, R.D.; Paza, A.O.; Matsumoto, M.A.N.; Proff, P.; Kirschneck, C. Potential interactions among single nucleotide polymorphisms in bone- and cartilage-related genes in skeletal malocclusions. Orthod. Craniofac. Res. 2021, 24, 277–287. [Google Scholar] [CrossRef]
- Nascimento, M.A.; Oliveira, D.S.B.; Reis, C.L.B.; Wambier, L.M.; Horta, K.C.; Romano, F.L.; Bezerra da Silva, L.A.; Raquel da Silva, A.B.; Nelson-Filho, P.; Kuchler, E.C. Association between P561T polymorphism in growth hormone receptor gene and mandibular prognathism: Systematic review and metaanalysis. Rev. Científica Do CRO-RJ 2019, 4, 2–11. [Google Scholar]
- Khundmiri, S.J.; Murray, R.; Lederer, E. PTH and Vitamin D. Compr. Physiol. 2016, 15, 561–601. [Google Scholar]
- Van der Velden, U.; Kuzmanova, D.; Chapple, I.L.C. Micronutritional approaches to periodontal therapy. J. Clin. Periodontol. 2011, 38, 142–158. [Google Scholar] [CrossRef]
- Beckerman, P.; Silver, J. Vitamin D and the parathyroid. Am. J. Med. Sci. 1999, 317, 363–369. [Google Scholar]
- Wolf, M.; Lossdörfer, S.; Marciniak, J.; Römer, P.; Kirschneck, C.; Craveiro, R.; Deschner, J.; Jäger, A. CD8+ T cells mediate the regenerative PTH effect in hPDL cells via Wnt10b signaling. Innate Immun. 2016, 22, 674–681. [Google Scholar] [CrossRef] [Green Version]
- Zhalehjoo, N.; Shakiba, Y.; Panjehpour, M. Gene expression profiles of CYP24A1 and CYP27B1 in malignant and normal breast tissues. Mol. Med. Rep. 2017, 15, 467–473. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.E.; Slice, D.E. Regional shape change in adult facial bone curvature with age. Am. J. Phys. Anthropol. 2010, 143, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, T.W.; Riminucci, M.; Holmbeck, K.; Bianco, P.; Robey, P.G. Development of craniofacial structures in transgenic mice with constitutively active PTH/PTHrP receptor. Bone 2008, 42, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Dutra, E.H.; O’Brien, M.H.; Gutierrez, T.; Lima, A.; Nanda, R.; Yadav, S. PTH [1-34]-induced alterations predispose the mandibular condylar cartilage to mineralization. Orthod. Craniofacial Res. 2017, 20, 162–166. [Google Scholar] [CrossRef]
- Engström, C.; Linde, A.; Thilander, B. Craniofacial morphology and growth in the rat. Cephalometric analysis of the effects of a low calcium and vitamin D-deficient diet. J. Anat. 1982, 134 Pt 2, 299–314. [Google Scholar]
- Little, J.; Higgins, J.P.; Ioannidis, J.P.; Moher, D.; Gagnon, F.; Von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement. Genet. Epidemiol. Off. Publ. Int. Genet. Epidemiol. Soc. 2009, 33, 581–598. [Google Scholar]
- Küchler, E.C.; Tannure, P.N.; Falagan-Lotsch, P.; Lopes, T.S.; Granjeiro, J.M.; Amorim, L.M.F. Buccal cells DNA extraction to obtain high quality human genomic DNA suitable for polymorphism genotyping by PCR-RFLP and Real-Time PCR. J. Appl. Oral Sci. 2012, 20, 467–471. [Google Scholar] [CrossRef] [Green Version]
- Cunha, A.S.; Dos Santos, L.V.; Marañón-Vásquez, G.A.; Kirschneck, C.; Gerber, J.T.; Stuani, M.B.; Matsumoto, M.A.N.; Vieira, A.R.; Scariot, R.; Küchler, E.C. Genetic variants in tooth agenesis–related genes might be also involved in tooth size variations. Clin. Oral Investig. 2021, 25, 1307–1318. [Google Scholar] [CrossRef]
- Ertl, D.A.; Stary, S.; Streubel, B.; Raimann, A.; Haeusler, G. A novel homozygous mutation in the parathyroid hormone gene (PTH) in a girl with isolated hypoparathyroidism. Bone 2012, 51, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Kanzawa, M.; Sugimoto, T.; Kobayashi, T.; Kobayashi, A.; Chihara, K. Parathyroid hormone gene polymorphisms in primary hyperparathyroidism. Clin. Endocrinol. 1999, 50, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Tenne, M.; McGuigan, F.; Jansson, L.; Gerdhem, P.; Obrant, K.J.; Luthman, H.; Åkesson, K. Genetic variation in the PTH pathway and bone phenotypes in elderly women: Evaluation of PTH, PTHLH, PTHR1 and PTHR2 genes. Bone 2008, 42, 719–727. [Google Scholar] [CrossRef]
- Al-Ghafari, A.B.; Balamash, K.S.; Al Doghaither, H.A. Relationship Between Serum Vitamin D and Calcium Levels and Vitamin D Receptor Gene Polymorphisms in Colorectal Cancer. BioMed Res. Int. 2019, 2019, 8571541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeljic, K.; Supic, G.; Stamenkovic, R.M.; Jovic, N.; Kozomara, R.; Magic, Z. Vitamin D receptor, CYP27B1 and CYP24A1 genes polymorphisms association with oral cancer risk and survival. J. Oral Pathol. Med. 2012, 41, 779–787. [Google Scholar] [CrossRef]
- Hibler, E.A.; Klimentidis, Y.C.; Jurutka, P.W.; Kohler, L.N.; Lance, P.; Roe, D.J.; Jacobs, E.T. CYP24A1 and CYP27B1 polymorphisms, concentrations of vitamin D metabolites, and odds of colorectal adenoma recurrence. Nutr. Cancer 2015, 67, 1131–1141. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Human Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Pattin, K.A.; White, B.C.; Barney, N.; Gui, J.; Nelson, H.H.; Kelsey, K.T.; Moore, J.H. A computationally efficient hypothesis testing method for epistasis analysis using multifactor dimensionality reduction. Genet. Epidemiol. Off. Publ. Int. Genet. Epidemiol. Soc. 2009, 33, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakulin, A.; Bratko, I. Quantifying and visualizing attribute interactions. arXiv 2004, arXiv:cs/0308002v3. [Google Scholar]
- Joshi, N.; Hamdan, A.M.; Fakhouri, W.D. Skeletal malocclusion: A developmental disorder with a life-long morbidity. J. Clin. Med. Res. 2014, 6, 399–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.R.; Burgess, J.A.; Critchlow, C.W. Association between mandibular retrognathia and TMJ disorders in adult females. J. Public Health Dent. 2004, 64, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Solow, B.; Siersbæk-Nielsen, S.; Greve, E. Airway adequacy, head posture, and craniofacial morphology. Am. J. Orthod. 1984, 86, 214–223. [Google Scholar] [CrossRef]
- Costa, A.M.G.; Trevizan, M.; Matsumoto, M.A.N.; da Silva, R.A.B.; da Silva, L.A.B.; Horta, K.C.; Romano, F.L.; Nelson-Filho, P.; Küchler, E.C. Association between tooth agenesis and skeletal malocclusions. J. Oral Maxillofac. Res. 2017, 8, e3. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, S.J.; Gomez, M.; Rey, J.A.; Ochoa, M.; Gutierrez, S.M.; Prieto, J.C. Polymorphisms of the noggin gene and mandibular micrognathia: A first approximation. Acta Odontol Lat. 2010, 23, 13–19. [Google Scholar]
- da Fontoura, C.G.; Miller, S.F.; Wehby, G.L.; Amendt, B.A.; Holton, N.E.; Southard, T.E.; Moreno Uribe, L.M. Candidate gene analyses of skeletal variation in malocclusion. J. Dent. Res. 2015, 94, 913–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barash, M.; Bayer, P.E.; van Daal, A. Identification of the Single Nucleotide Polymorphisms Affecting Normal Phenotypic Variability in Human Craniofacial Morphology Using Candidate Gene Approach. J. Genet. Genome Res. 2018, 5, 041. [Google Scholar]
- Barros, S.P.; Silva, M.A.D.; Somerman, M.J.; Nociti, F.H., Jr. Parathyroid hormone protects against periodontitis-associated bone loss. J. Dent. Res. 2013, 82, 791–795. [Google Scholar] [CrossRef]
- Tokunaga, K.; Seto, H.; Ohba, H.; Mihara, C.; Hama, H.; Horibe, M.; Yoneda, S.; Nagata, T. Topical and intermittent application of parathyroid hormone recovers alveolar bone loss in rat experimental periodontitis. J. Periodontal Res. 2011, 46, 655–662. [Google Scholar] [CrossRef]
- Silva, B.C.; Costa, A.G.; Cusano, N.E.; Kousteni, S.; Bilexikian, J.P. Catabolic and anabolic actions of parathyroid hormone on the skeleton. J. Endocrinol. Investig. 2011, 34, 801–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Hargreaves, K.A.; Li, R.; Reiter, J.L.; Wang, Y.; Mort, M.; Cooper, D.N.; Zhou, Y.; Zhang, C.; Eadon, M.T.; et al. RegSNPs-intron: A computational framework for predicting pathogenic impact of intronic single nucleotide variants. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Hughes, T.A. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006, 22, 119–122. [Google Scholar] [CrossRef]
- Lehmann, B.; Meurer, M. Vitamin D metabolism. Dermatol. Ther. 2010, 23, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, K.; Abe, T.; Oue, N.; Yasui, W.; Ryoji, M. Characterization of vitamin D-mediated induction of the CYP 24 transcription. Mol. Cell. Endocrinol. 2004, 226, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Segersten, U.; Björklund, P.; Hellman, P.; Åkerström, G.; Westin, G. Potentiating effects of nonactive/active vitamin D analogues and ketoconazole in parathyroid cells. Clin. Endocrinol. 2007, 66, 399–404. [Google Scholar] [CrossRef] [PubMed]



| Gene | SNP | Base Change # | Functional Consequence # | Clinical Significance †,‡ | Biological Effects (Reference) |
|---|---|---|---|---|---|
| PTH | rs694 | C > T | Intron Variant | Benign | Low levels of PTH serum [25] |
| rs6256 * | G > T | Stop Gained * | Benign | Low levels of PTH serum [26] | |
| rs307247 | G > A | 3′ untranslated region | Benign | Low levels of PTH serum [27] | |
| VDR | rs7975232 # | C > A | Intron Variant | Benign | High levels of Vitamin-D serum [28] |
| CYP27B1 | rs464653 | A > G | Intron Variant | Benign | Oral Neoplasm [29] |
| CYP24A1 | rs927650 | C > T | Intron Variant | Uncertain Significance | High levels of Vitamin-D serum [30] |
| Variables | MR | Control | p-Value |
|---|---|---|---|
| Gender n (%) | |||
| Male | 22 (45.8) | 17 (39.5) | 0.672 |
| Female | 26 (54.2) | 26 (60.1) | |
| Age | |||
| Median (IQR) | 12.0 (4.0) | 12.0 (4.5) | 0.809 |
| SNB (°) | |||
| Median (IQR) | 76.1 (3.5) | 80.0 (2.0) | <0.001 * |
| ANB (°) | |||
| Mean (SD) | 4.2 (2.3) | 2.4 (2.3) | <0.001 * |
| Co-Gn (mm) | |||
| Mean (SD) | 110 (8.0) | 115 (9.4) | 0.008 * |
| Go-Pg (mm) | |||
| Median (IQR) | 64.4 (6.9) | 68.2 (5.5) | 0.014 * |
| Co-Go (mm) | |||
| Median (IQR) | 54.0 (8.8) | 56.8 (7.4) | 0.044 * |
| Gene | SNP | Frequency-n (%) | p-Value | OR (CI 95%) | |||
|---|---|---|---|---|---|---|---|
| Genotype/Allele | Control | MR | |||||
| PTH | rs694 | Genotype | TT | 4 (10.8) | 19 (44.2) | Reference | - |
| CT | 22 (59.5) | 19 (44.2) | 0.004 * | 0.18 (0.06–0.59) | |||
| CC | 11 (29.7) | 5 (11.6) | 0.001 * | 0.09 (0.02–0.44) | |||
| Allele | T | 30 (40.5) | 57 (59.4) | Reference | - | ||
| C | 44 (59.5) | 39 (40.6) | 0.014 * | 0.46 (0.25–0.87) | |||
| rs6256 | Genotype | GG | 30 (81.1) | 32 (76.2) | Reference | ||
| GT | 7 (18.9) | 9 (21.4) | 0.740 | 1.20 (0.39–3.39) | |||
| TT | 0 (0.0) | 1 (2.4) | >0.999 | - | |||
| Allele | G | 67 (90.5) | 73 (86.9) | Reference | - | ||
| T | 7 (9.5) | 11 (13.1) | 0.472 | 1.44 (0.51–3.71) | |||
| rs307247 | Genotype | GG | 12 (33.3) | 26 (60.5) | Reference | ||
| AG | 14 (38.9) | 12 (27.9) | 0.074 | 0.39 (0.14–1.16) | |||
| AA | 10 (27.8) | 5 (11.6) | 0.019 * | 0.23 (0.06–0.78) | |||
| Allele | G | 38 (52.8) | 64 (75.0) | Reference | |||
| A | 34 (47.2) | 22 (25.0) | 0.003 * | 0.37 (0.19–0.73) | |||
| VDR | rs7975232 | Genotype | AA | 14 (35.0) | 18 (41.9) | Reference | |
| AC | 17 (42.5) | 21 (48.8) | 0.934 | 0.96 (0.37–2.44) | |||
| CC | 9 (22.5) | 4 (9.3) | 0.121 | 0.34 (0.10–1.31) | |||
| Allele | A | 45 (56.2) | 57 (66.3) | Reference | - | ||
| C | 35 (43.8) | 29 (33.7) | 0.184 | 0.65 (0.34–1.20) | |||
| CYP27B1 | rs464653 | Genotype | AA | 13 (33.3) | 25 (55.6) | Reference | - |
| AG | 21 (53.9) | 15 (33.3) | 0.037 * | 0.37 (0.13–0.93) | |||
| GG | 5 (12.8) | 5 (11.1) | 0.358 | 0.52 (0.12–2.09) | |||
| Allele | A | 47 (60.3) | 45 (64.3) | Reference | - | ||
| G | 31 (39.7) | 25 (35.7) | 0.613 | 0.84 (0.44–1.68) | |||
| CYP24A1 | rs927650 | Genotype | CC | 17 (44.7) | 15 (34.1) | Reference | - |
| CT | 16 (42.1) | 26 (59.1) | 0.197 | 1.84 (0.75–4.69) | |||
| TT | 5 (13.2) | 3 (6.8) | 0.633 | 0.68 (0.16–3.45) | |||
| Allele | C | 50 (65.8) | 56 (63.6) | Reference | - | ||
| T | 26 (34.2) | 32 (36.4) | 0.773 | 1.09 (0.57–2.12) | |||
| Locus Number | Best Combination | CVC # | TBA † | p-Value ‡ |
|---|---|---|---|---|
| 2 | rs694 (PTH), rs927650 (CYP24A1) | 10/10 | 0.6742 | 0.040 * |
| 3 | rs307247 (PTH), rs464653 (CYP27B1), rs927650 (CYP24A1) | 10/10 | 0.7651 | <0.001 * |
| 4 | rs307247 (PTH), rs7975232 (VDR), rs464653 (CYP27B1), rs927650 (CYP24A1) | 8/10 | 0.7016 | 0.009 * |
| 5 | rs694 (PTH), rs307247 (PTH), rs7975232 (VDR), rs464653 (CYP27B1), rs927650 (CYP24A1) | 8/10 | 0.6832 | 0.026 * |
| 6 | rs694 (PTH), rs6256 (PTH), rs307247 (PTH), rs7975232 (VDR), rs464653 (CYP27B1), rs927650 (CYP24A1) | 10/10 | 0.7085 | 0.008 * |
| SNPS | Haplotype | MR | Control | p-Value |
|---|---|---|---|---|
| rs694, rs6256, rs307247 | C-G-A | 0.25 | 0.47 | 0.004 * |
| C-T-G | 0.01 | 0.03 | 0.510 | |
| T-T-G | 0.11 | 0.06 | 0.231 | |
| C-G-G | 0.06 | 0.09 | 0.530 | |
| T-G-G | 0.54 | 0.34 | 0.010 * | |
| rs694, rs6256 | T-T | 0.13 | 0.09 | 0.472 |
| C-G | 0.33 | 0.59 | 0.001 * | |
| T-G | 0.53 | 0.31 | 0.004 * | |
| rs694, rs307247 | C-A | 0.25 | 0.47 | 0.004 * |
| C-G | 0.08 | 0.12 | 0.365 | |
| T-G | 0.66 | 0.40 | 0.001 * | |
| rs307247, rs6256 | A-G | 0.26 | 0.47 | 0.006 * |
| G-T | 0.13 | 0.09 | 0.510 | |
| G-G | 0.60 | 0.43 | 0.027 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Küchler, E.C.; Reis, C.L.B.; Marañón-Vásquez, G.; Nelson-Filho, P.; Matsumoto, M.A.N.; Stuani, M.B.S.; Oliveira, M.A.H.d.M.; Proff, P.; Kirschneck, C. Parathyroid Hormone Gene and Genes Involved in the Maintenance of Vitamin D Levels Association with Mandibular Retrognathism. J. Pers. Med. 2021, 11, 369. https://doi.org/10.3390/jpm11050369
Küchler EC, Reis CLB, Marañón-Vásquez G, Nelson-Filho P, Matsumoto MAN, Stuani MBS, Oliveira MAHdM, Proff P, Kirschneck C. Parathyroid Hormone Gene and Genes Involved in the Maintenance of Vitamin D Levels Association with Mandibular Retrognathism. Journal of Personalized Medicine. 2021; 11(5):369. https://doi.org/10.3390/jpm11050369
Chicago/Turabian StyleKüchler, Erika Calvano, Caio Luiz Bitencourt Reis, Guido Marañón-Vásquez, Paulo Nelson-Filho, Mírian Aiko Nakane Matsumoto, Maria Bernadete Sasso Stuani, Maria Angélica Hueb de Menezes Oliveira, Peter Proff, and Christian Kirschneck. 2021. "Parathyroid Hormone Gene and Genes Involved in the Maintenance of Vitamin D Levels Association with Mandibular Retrognathism" Journal of Personalized Medicine 11, no. 5: 369. https://doi.org/10.3390/jpm11050369
APA StyleKüchler, E. C., Reis, C. L. B., Marañón-Vásquez, G., Nelson-Filho, P., Matsumoto, M. A. N., Stuani, M. B. S., Oliveira, M. A. H. d. M., Proff, P., & Kirschneck, C. (2021). Parathyroid Hormone Gene and Genes Involved in the Maintenance of Vitamin D Levels Association with Mandibular Retrognathism. Journal of Personalized Medicine, 11(5), 369. https://doi.org/10.3390/jpm11050369