Pharmacogenetic Profiling in High-Risk Soft Tissue Sarcomas Treated with Neoadjuvant Chemotherapy
Abstract
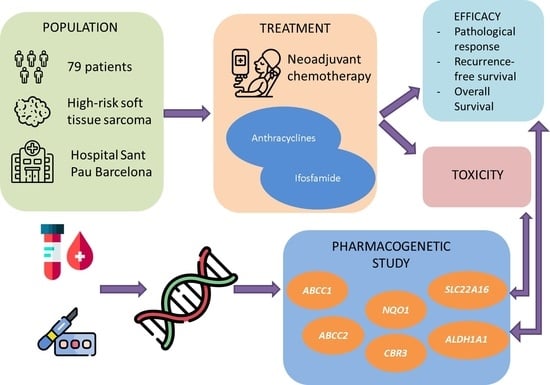
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping
2.3. Statistics
3. Results
3.1. Genetic Variants and Toxicity
3.2. Genetic Variants and Survival
3.3. Genetic Variants and Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours WHO Classification of Tumours, 5th ed.; IARC: Lyon, France, 2020; Volume 3. [Google Scholar]
- Casali, P.G.; Abecassis, N.; Bauer, S.; Biagini, R.; Bielack, S.; Bonvalot, S.; Boukovinas, I.; Bovee, J.V.M.G.; Brodowicz, T.; Broto, J.; et al. Soft tissue and visceral sarcomas: ESMO–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29 (Suppl. 4), iv51–iv67. [Google Scholar] [CrossRef] [PubMed]
- Baldini, E.H.; Le Cesne, A.; Trent, J.C. Neoadjuvant Chemotherapy, Concurrent Chemoradiation, and Adjuvant Chemotherapy for High-Risk Extremity Soft Tissue Sarcoma. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharm. Genom. 2011, 21, 440. Available online: http://www.pharmgkb.org/do/serve?objId (accessed on 5 February 2019). [CrossRef] [PubMed]
- Gewirtz, D. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar] [CrossRef]
- Lal, S.; Mahajan, A.; Chen, W.N.; Chowbay, B. Pharmacogenetics of Target Genes across Doxorubicin Disposition Pathway: A Review. Curr. Drug Metab. 2010, 11, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Fung, K.L.; Gottesman, M.M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2009, 1794, 860–871. [Google Scholar] [CrossRef] [Green Version]
- Okabe, M.; Unno, M.; Harigae, H.; Kaku, M.; Okitsu, Y.; Sasaki, T.; Mizoi, T.; Shiiba, K.; Takanaga, H.; Terasaki, T.; et al. Characterization of the organic cation transporter SLC22A16: A doxorubicin importer. Biochem. Biophys. Res. Commun. 2005, 333, 754–762. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0006291X05011873 (accessed on 21 May 2019). [CrossRef]
- Fagerholm, R.; Hofstetter, B.; Tommiska, J.; Aaltonen, K.; Vrtel, R.; Syrjäkoski, K.; Kallioniemi, A.; Kilpivaara, O.; Mannermaa, A.; Kosma, V.-M.; et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet. 2008, 40, 844–853. Available online: https://www.nature.com/articles/ng.155 (accessed on 30 July 2008). [CrossRef]
- Fan, L.; Goh, B.-C.; Wong, C.-I.; Sukri, N.; Lim, S.-E.; Tan, S.-H.; Guo, J.-Y.; Lim, R.; Yap, H.-L.; Khoo, Y.-M.; et al. Genotype of human carbonyl reductase CBR3 correlates with doxorubicin disposition and toxicity. Pharm. Genom. 2008, 18, 623–631. Available online: http://www.bioconductor.org (accessed on 4 December 2018). [CrossRef]
- Lowenberg, D.; Thorn, C.F.; Desta, Z.; Flockhart, D.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Ifosfamide pathways, pharmacokinetics and pharmacodynamics. Pharm. Genom. 2014, 24, 133. Available online: http://www.pharmgkb.org/pathway/PA2037 (accessed on 5 February 2019). [CrossRef] [Green Version]
- Evans, W.E.; McLeod, H.L. Pharmacogenomics—Drug Disposition, Drug Targets, and Side Effects. N. Engl. J. Med. 2003, 348, 538–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tęcza, K.; Pamuła-Piłat, J.; Lanuszewska, J.; Butkiewicz, D.; Grzybowska, E. Pharmacogenetics of toxicity of 5-fluorouracil, doxorubicin and cyclophosphamide chemotherapy in breast cancer patients. Oncotarget 2018, 9, 9114–9136. [Google Scholar] [CrossRef] [Green Version]
- Lal, S.; Wong, Z.W.; Sandanaraj, E.; Xiang, X.; Ang, P.C.S.; Lee, E.J.D.; Chowbay, B. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008, 99, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Bray, J.; Sludden, J.; Griffin, M.J.; Cole, M.; Verrill, M.; Jamieson, D.; Boddy, A.V. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br. J. Cancer 2010, 102, 1003–1009. [Google Scholar] [CrossRef] [Green Version]
- Geng, R.; Chen, Z.; Zhao, X.; Qiu, L.; Liu, X.; Liu, R.; Guo, W.; He, G.; Li, J.; Zhu, X. Oxidative Stress-Related Genetic Polymorphisms Are Associated with the Prognosis of Metastatic Gastric Cancer Patients Treated with Epirubicin, Oxaliplatin and 5-Fluorouracil Combination Chemotherapy. PLoS ONE 2014, 9, e116027. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Sucheston, L.E.; Zhao, H.; Barlow, W.E.; Zirpoli, G.; Liu, S.; Moore, H.C.F.; Budd, G.T.; Hershman, D.L.; Davis, W.; et al. Germline genetic variants in ABCB1, ABCC1 and ALDH1A1, and risk of hematological and gastrointestinal toxicities in a SWOG Phase III trial S0221 for breast cancer. Pharm. J. 2013, 14, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. 2009. Available online: http://www.meddramsso.com (accessed on 19 April 2021).
- Wardelmann, E.; Haas, R.L.; Bovée, J.V.M.G.; Terrier, P.; Lazar, A.; Messiou, C.; Lepechoux, C.; Hartmann, W.; Collin, F.; Fisher, C.; et al. Evaluation of response after neoadjuvant treatment in soft tissue sarcomas; the European Organization for Research and Treatment of CancereSoft Tissue and Bone Sarcoma Group (EORTCeSTBSG) recommendations for pathological examination and reporting. Eur. J. Cancer 2016, 53, 84–95. [Google Scholar] [CrossRef] [PubMed]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Gregers, J.; Gréen, H.; Christensen, I.J.; Dalhoff, K.; Schroeder, H.; Carlsen, N.; Rosthoej, S.; Lausen, B.; Schmiegelow, K.; Peterson, C. Polymorphisms in the ABCB1 gene and effect on outcome and toxicity in childhood acute lymphoblastic leukemia. Pharm. J. 2015, 15, 372–379. [Google Scholar] [CrossRef]
- Ikeda, M.; Tsuji, D.; Yamamoto, K.; Kim, Y.-I.; Daimon, T.; Iwabe, Y.; Hatori, M.; Makuta, R.; Hayashi, H.; Inoue, K.; et al. Relationship between ABCB1 gene polymorphisms and severe neutropenia in patients with breast cancer treated with doxorubicin/cyclophosphamide chemotherapy. Drug Metab. Pharmacokinet. 2015, 30, 149–153. [Google Scholar] [CrossRef]
- Hertz, D.L.; Caram, M.V.; Kidwell, K.M.; Thibert, J.N.; Gersch, C.; Seewald, N.J.; Smerage, J.; Rubenfire, M.; Henry, N.L.; Cooney, K.A.; et al. Evidence for association of SNPs in ABCB1 and CBR3, but not RAC2, NCF4, SLC28A3 or TOP2B, with chronic cardiotoxicity in a cohort of breast cancer patients treated with anthracyclines. Pharmacogenomics 2016, 17, 231–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggini, V.; Buda, G.; Martino, A.; Presciuttini, S.; Galimberti, S.; Orciuolo, E.; Barale, R.; Petrini, M.; Rossi, A.M. MDR1 diplotypes as prognostic markers in multiple myeloma. Pharm. Genom. 2008, 18, 283–289. Available online: https://insights.ovid.com/pubmed?pmid=18408561 (accessed on 5 February 2019). [CrossRef] [PubMed]
- Cizmarikova, M.; Wagnerova, M.; Schonova, L.; Habalova, V.; Kohut, A.; Linkova, A.; Sarissky, M.; Mojzis, J.; Mirossay, L. MDR1 (C3435T) polymorphism: Relation to the risk of breast cancer and therapeutic outcome. Pharm. J. 2009, 10, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Caronia, D.; Patiño-García, A.; Martínez, A.P.; Pita, G.; Moreno, L.T.; Zalacain-Díez, M.; Molina, B.; Colmenero, I.; Sierrasesúmaga, L.; Benítez, J.F.; et al. Effect of ABCB1 and ABCC3 Polymorphisms on Osteosarcoma Survival after Chemotherapy: A Pharmacogenetic Study. PLoS ONE 2011, 6, e26091. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.-I.; Nagashima, F.; Yamamoto, W.; Endo, H.; Sunakawa, Y.; Yamashita, K.; Ishida, H.; Mizuno, K.; Matsunaga, M.; Araki, K.; et al. Association of ATP-Binding Cassette, Sub-family C, Number 2 (ABCC2) Genotype with Pharmacokinetics of Irinotecan in Japanese Patients with Metastatic Colorectal Cancer Treated with Irinotecan Plus Infusional 5-Fluorouracil/Leucovorin (FOLFIRI). Biol. Pharm. Bull. 2008, 31, 2137–2142. [Google Scholar] [CrossRef] [Green Version]
- Innocenti, F.; Kroetz, D.L.; Schuetz, E.; Dolan, M.E.; Ramírez, J.; Relling, M.; Chen, P.; Das, S.; Rosner, G.L.; Ratain, M.J. Comprehensive Pharmacogenetic Analysis of Irinotecan Neutropenia and Pharmacokinetics. J. Clin. Oncol. 2009, 27, 2604–2614. [Google Scholar] [CrossRef] [Green Version]
- Ravegnini, G.; Urbini, M.; Simeon, V.; Genovese, C.; Astolfi, A.; Nannini, M.; Gatto, L.; Saponara, M.; Ianni, M.; Indio, V.; et al. An exploratory study by DMET array identifies a germline signature associated with imatinib response in gastrointestinal stromal tumor. Pharm. J. 2018, 19, 390–400. [Google Scholar] [CrossRef]
- Goričar, K.; Kovač, V.; Dolžan, V. Clinical-pharmacogenetic models for personalized cancer treatment: Application to malignant mesothelioma. Sci. Rep. 2017, 7, srep46537. [Google Scholar] [CrossRef] [Green Version]
- Haenisch, S.; May, K.; Wegner, D.; Caliebe, A.; Cascorbi, I.; Siegmund, W. Influence of genetic polymorphisms on intestinal expression and rifampicin-type induction of ABCC2 and on bioavailability of talinolol. Pharm. Genom. 2008, 18, 357–365. [Google Scholar] [CrossRef]
- Lian, G.; Yuan, J.; Gao, Y. In vitro Transport Ability of ABCC2 (G1249A) Polymorphic Variant towards Anticancer Drugs. OncoTargets Ther. 2020, 13, 1413–1419. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, D.; Cresti, N.; Bray, J.; Sludden, J.; Griffin, M.J.; Hawsawi, N.M.; Famie, E.; Mould, E.V.; Verrill, M.W.; May, F.E.; et al. Two minor NQO1 and NQO2 alleles predict poor response of breast cancer patients to adjuvant doxorubicin and cyclophosphamide therapy. Pharm. Genom. 2011, 21, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Serie, D.J.; Crook, J.E.; Necela, B.M.; Dockter, T.J.; Wang, X.; Asmann, Y.W.; Fairweather, D.; Bruno, K.A.; Colon-Otero, G.; Perez, E.A.; et al. Genome-wide association study of cardiotoxicity in the NCCTG N9831 (Alliance) adjuvant trastuzumab trial. Pharm. Genom. 2017, 27, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riera, P.; Artigas-Baleri, A.; Salazar, J.; Sebio, A.; Virgili, A.C.; Arranz, M.J.; Páez, D. ABCB1 Genetic Variants as Predictors of Irinotecan-Induced Severe Gastrointestinal Toxicity in Metastatic Colorectal Cancer Patients. Front. Pharmacol. 2020, 11, 973. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, D.; Boddy, A.V. Pharmacogenetics of genes across the doxorubicin pathway. Expert Opin. Drug Metab. Toxicol. 2011, 7, 1201–1210. [Google Scholar] [CrossRef]
- Chan, L.M.; Lowes, S.; Hirst, B.H. The ABCs of drug transport in intestine and liver: Efflux proteins limiting drug absorption and bioavailability. Eur. J. Pharm. Sci. 2004, 21, 25–51. [Google Scholar] [CrossRef]
- Haenisch, S.; Zimmermann, U.; Dazert, E.; Wruck, C.J.; Dazert, P.; Siegmund, S.; Kroemer, H.K.; Warzok, R.W.; Cascorbi, I. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharm. J. 2006, 7, 56–65. [Google Scholar] [CrossRef]
- Vasiliou, V.; Pappa, A. Polymorphisms of human aldehyde dehydrogenases: Consequences for drug metabolism and disease. Pharmacology 2000, 61, 192–198. Available online: https://www.karger.com/Article/FullText/28400 (accessed on 31 May 2021). [CrossRef]
- Liu, J.; Zhou, Z.; Hodgkinson, C.A.; Yuan, Q.; Shen, P.-H.; Mulligan, C.J.; Wang, A.; Gray, R.R.; Roy, A.; Virkkunen, M.; et al. Haplotype-Based Study of the Association of Alcohol-Metabolizing Genes with Alcohol Dependence in Four Independent Populations. Alcohol. Clin. Exp. Res. 2010, 35, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Bunting, K.D.; Townsend, A.J. De novo expression of transfected human class 1 aldehyde dehydrogenase (ALDH) causes resistance to oxazaphosphorine anti-cancer alkylating agents in hamster V79 cell lines. Elevated class 1 ALDH activity is closely correlated with reduction in DNA interstrand cross-linking and lethality. J. Biol. Chem. 1996, 271, 11884–11890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreb, J.S.; Schweder, M.; Gray, B.; Zucali, J.; Zori, R. In VitroSelection for K562 Cells with Higher Retrovirally Mediated Copy Number of Aldehyde Dehydrogenase Class-1 and Higher Resistance to 4-Hydroperoxycyclophosphamide. Hum. Gene Ther. 1998, 9, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Canuto, R.A.; Muzio, G.; Salvo, R.A.; Maggiora, M.; Trombetta, A.; Chantepie, J.; Fournet, G.; Reichert, U.; Quash, G. The effect of a novel irreversible inhibitor of aldehyde dehydrogenases 1 and 3 on tumour cell growth and death. Chem. Interact. 2001, 130–132, 209–218. [Google Scholar] [CrossRef]
- Khoury, T.; Ademuyiwa, F.; Chandraseekhar, R.; Jabbour, M.; DeLeo, A.B.; Ferrone, S.; Wang, Y.; Wang, X. Aldehyde dehydrogenase 1A1 expression in breast cancer is associated with stage, triple negativity, and outcome to neoadjuvant chemotherapy. Mod. Pathol. 2011, 25, 388–397. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.-H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef] [Green Version]
- McWhinney, S.R.; McLeod, H.L. Using germline genotype in cancer pharmacogenetic studies. Pharmacogenomics 2009, 10, 489–493. Available online: https://pubmed.ncbi.nlm.nih.gov/19650256/ (accessed on 25 August 2021). [CrossRef] [Green Version]

| Patient and Tumour Characteristics (n = 95) | n | % |
|---|---|---|
| Age (years) | ||
| Median | 53 | |
| Range | 19–77 | |
| <60 | 68 | 71.6 |
| ≥60 | 27 | 28.4 |
| Sex | ||
| Male | 59 | 62.1 |
| Female | 36 | 37.9 |
| ECOG * performance status | ||
| 0 | 34 | 35.8 |
| 1 | 40 | 42.1 |
| 2 | 4 | 4.2 |
| Unknown | 17 | 17.9 |
| Histology | ||
| Undifferentiated pleomorphic sarcoma | 28 | 29.5 |
| Synovial sarcoma | 19 | 20.0 |
| Spindle cell sarcoma, NOS ** | 15 | 15.8 |
| Leiomyosarcoma | 10 | 10.5 |
| Myxofibrosarcoma | 7 | 7.4 |
| Myxoid liposarcoma | 3 | 3.2 |
| Pleomorphic liposarcoma | 3 | 3.2 |
| Malignant peripheral nerve sheath tumour | 3 | 3.2 |
| Others | 7 | 7.4 |
| Site | ||
| Lower limb | 73 | 76.8 |
| Upper limb | 16 | 16.8 |
| Trunk | 6 | 6.3 |
| Chemotherapy | ||
| Epirubicin-ifosfamide | 61 | 64.2 |
| High-dose ifosfamide | 24 | 25.2 |
| Others | 10 | 10.5 |
| Radiotherapy | ||
| Neoadjuvant | 40 | 42.1 |
| Adjuvant | 36 | 37.9 |
| Neoadjuvant and adjuvant | 12 | 12.6 |
| No | 7 | 7.4 |
| Pathological response | ||
| ≥90% | 35 | 36.8 |
| <90% | 44 | 56.8 |
| Not evaluable | 10 | 6.3 |
| Drug Pathway/Gene Symbol | refSeq | MAF (Minor Allele) | SNP Label | Protein Label | References for Rationale |
|---|---|---|---|---|---|
| ANTHRACYCLINES | |||||
| ABCB1 | rs1045642 | 0.48 (C) | c.3435T>C | p.Ile1145= | [7,14,21,22,23,24,25] |
| rs2032582 | 0.41 (T); 0.02 (A) | c.2677T>G; c.2677T>A | p.Ser893Ala; p.Ser893Thr | [14,15,22,24] | |
| rs1128503 | 0.42 (T) | c.1236T>C | p.Gly412= | [14,26] | |
| ABCC2 | rs3740066 | 0.37 (T) | c.3972C>T | p.Ile1324= | [13,27,28] |
| rs2273697 | 0.20 (A) | c.1249G>A | p.Val417Ile | [13,27,29,30,31,32] | |
| NQO1 | rs1800566 | 0.21 (T) | c.559C>T | p.Pro187Ser | [9,16,33] |
| CBR3 | rs8133052 | 0.45 (A) | c.11G>A | p.Cys4Tyr | [10] |
| rs1056892 | 0.35 (A) | c.730G>A | p.Val244Met | [10,23,34] | |
| SLC22A16 | rs6907567 * | 0.22 (C) | c.312T>C | p.Asn104= | [29,31] |
| rs12210538 | 0.24 (C) | c.1226T>C | p.Met409Thr | [15] | |
| IFOSFAMIDE | |||||
| ALDH1A1 | rs3764435 | 0.49 (G) | c.1434-680T>G | [17] | |
| rs168351 | 0.16 (C) | c.1434-1115T>C | [17] |
| n | G3-4 Anaemia n (%) | G3-4 Thrombo- Cytopenia n (%) | G3-4 Neutropenia n (%) | Febrile Neutropenia n (%) | G3-4 Transaminitis n (%) | Haemorrhagic Cystitis n (%) | Pathological Response > 90% n (%) | |
|---|---|---|---|---|---|---|---|---|
| ANTHRACYCLINES | ||||||||
| ABCC1—rs1045642 | ||||||||
| GG | 19 | 5 (26.3%) | 1 (5.3%) | 8 (42.1%) | 7 (36.8%) | 2 (10.5%) | 5 (29.4%) | |
| AG | 27 | 7 (25.9%) | 7 (25.9%) | 17 (63%) | 11 (40.7%) | 1 (3.8%) | 10 (41.7%) | |
| AA | 7 | 2 (28.6%) | 1 (14.3%) | 3 (42.9%) | 3 (42.9%) | 0 (0%) | 1 (14.3%) | |
| p-value | 1 * | 0.22 * | 0.323 * | 1 * | 0.721 * | 0.413 * | ||
| ABCC1—rs2032582 | ||||||||
| CC | 25 | 8 (32%) | 2 (8%) | 13 (52%) | 10 (40%) | 2 (8%) | 9 (42.9%) | |
| CT/CA | 23 | 4 (17.4%) | 6 (26.1%) | 12 (52.2%) | 8 (34.8%) | 1 (4.5%) | 6 (27.3%) | |
| TT/TA | 5 | 1 (20%) | 0 (0%) | 2 (40%) | 2 (40%) | 0 (0%) | 0 (0%) | |
| p-value | 0.528 * | 0.173 * | 1 * | 0.917 * | 1 * | 0.192 * | ||
| ABCC1—rs1128503 | ||||||||
| GG | 24 | 7 (29.2%) | 3 (12.5%) | 12 (50%) | 10 (41.7%) | 2 (8.3%) | 7 (33.3%) | |
| AG | 24 | 5 (20.8%) | 5 (20.8%) | 13 (54.2%) | 8 (33.3%) | 1 (4.3%) | 8 (36.4% | |
| AA | 6 | 2 (33.3%) | 1 (16.7%) | 3 (50%) | 3 (50%) | 0 (0%) | 1 (16.7%) | |
| p-value | 0.744 | 0.873 * | 1 * | 0.786 * | 1 * | 0.765 * | ||
| ABCC2—rs3740066 | ||||||||
| CC | 18 | 2 (11.21%) | 2 (11.1%) | 7 (38.7%) | 5 (27.8%) | 0 (0%) | 5 (31.3%) | |
| CT | 24 | 7 (29.2%) | 4 (16.7%) | 13 (54.2%) | 8 (33.3%) | 2 (8.7%) | 8 (36.4%) | |
| TT | 9 | 4 (44.4%) | 2 (22.2%) | 7 (77.8%) | 7 (77.8%) | 1 (11.1%) | 3 (37.5%) | |
| p-value | 0.167 * | 0.784 * | 0.179 * | 0.04 * | 0.398 | 1.000 * | ||
| ABCC2—rs2273697 | ||||||||
| GG | 35 | 12 (34.3%) | 8 (22.9%) | 20 (57.1%) | 17 (48.6%) | 2 (5.9%) | 13 (39.4%) | |
| AG | 15 | 2 (13.3%) | 1 (6.7%) | 6 (40%) | 4 (26.7%) | 1 (6.7%) | 2 (15.4%) | |
| AA | 4 | 0 (0%) | 0 (0%) | 2 (50%) | 0 (0%) | 0 (0%) | 1 (33.3%) | |
| p-value | 0.167 * | 0.330 * | 0.571 * | 0.103 * | 1 * | 0.219 * | ||
| NQO1— rs1800566 | ||||||||
| GG | 34 | 10 (29.4%) | 7 (20.6%) | 21 (61.8%) | 15 (44.1%) | 2 (6.1%) | 11 (35.5%) | |
| AG | 16 | 3 (18.8%) | 1 (6.3%) | 4 (25%) | 3 (18.8%) | 1 (6.3%) | 2 (14.3%) | |
| AA | 4 | 1 (25%) | 1 (25%) | 3 (75%) | 3 (75%) | 0(0%) | 3 (75.0%) | |
| p-value | 0.785 * | 0.403 * | >0.028 * | 0.058 * | 0.059 * | |||
| CBR3—rs1056892 | ||||||||
| GG | 30 | 9 (30%) | 5 (16.7%) | 17 (56.7%) | 13 (43.3%) | 2 (6.9%) | 12 (42.9%) | |
| AG | 18 | 4 (22.2%) | 3 (16.7%) | 8 (44.4%) | 5 (27.8%) | 1 (5.6%) | 1 (6.7%) | |
| AA | 6 | 1 (16.7%) | 1 (16.7%) | 3 (50%) | 3 (50%) | 0 (0%) | 5 (50%) | |
| p-value | 0.825 * | 1 * | 0.792 * | 0.479 * | 1 * | 0.024 * | ||
| SLC22A16—rs6907567 | ||||||||
| AA | 29 | 9 (31%) | 5 (17.2%) | 16 (55.2%) | 12 (41.4%) | 1 (3.6%) | 11 (42.3%) | |
| AG | 18 | 3 (16.7%) | 4 (22.2%) | 10 (55.6%) | 7 (38.9%) | 2 (11.1%) | 3 (18.8%) | |
| GG | 7 | 2 (28.6%) | 0 (0%) | 2 (28.6%) | 2 (28.6%) | 0 (0%) | 2 (28.6%) | |
| p-value | 0.564 * | 0.460 * | 0.478 | 0.926 | 0.71 | 0.256 * | ||
| IFOSFAMIDE | ||||||||
| ALDH1A1—rs3764435 | ||||||||
| AA | 23 | 5 (21.7%) | 2 (8.7%) | 9 (39.1%) | 6 (26.1%) | 3 (13.6%) | 1 (4.5%) | 9 (40.9%) |
| AC | 30 | 8 (26.7%) | 5 (16.7%) | 15 (50%) | 11 (36.9%) | 0 (0%) | 1 (3.3%) | 10 (38.5%) |
| CC | 18 | 3 (16.7%) | 1 (5.6%) | 7 (38.9%) | 7 (38.9%) | 0 (0%) | 0 (0%) | 6 (35.3%) |
| p-value | 0.771 * | 0.636 | 0.713 * | 0.657 * | 1 * | 0.949 * | ||
| ALDH1A1—rs168351 | ||||||||
| AA | 56 | 14 (25%) | 7 (12.5%) | 24 (42.9%) | 19 (33.9%) | 2 (3.6%) | 1 (1.8%) | 17 (33.3%) |
| AG | 14 | 1 (7.1%) | 1 (7.1%) | 6 (42.9%) | 4 (28.6%) | 0 (0%) | 1 (7.7%) | 8 (61.5%) |
| GG | 2 | 1 (50%) | 1 (50%) | 2 (100%) | 1 (50%) | 1 (50%) | 0 (0%) | 0 (0%) |
| p-value | 0.186 * | 0.291 * | 0.377 * | 1 * | 0.38 * | 0.097 * | ||
| SNP | n | OS | RFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Probability ± s.e * at 3-y | Probability ± s.e at 5-y | HR (95% CI) | p-Value | Probability ± s.e at 3-y | Probability ± s.e at 5-y | HR (95% CI) | p-Value | ||
| ANTHRACYCLINES | |||||||||
| ABCB1—rs1045642 | |||||||||
| GG | 16 | 0.83 ± 0.11 | 0.68 ± 0.17 | 1 (reference) | 0.352 | 0.47 ± 0.13 | 0.47 ± 0.13 | 1 (reference) | 0.712 |
| AG | 26 | 0.73 ± 0.10 | 0.61 ± 0.11 | 1.57 (0.42–5.85) | 0.48 ± 0.11 | 0.48 ± 0.11 | 1.01 (0.42–2.45) | ||
| AA | 7 | 1.00 ± 0.00 | 0.80 ± 0.18 | 0.41 (1.04–3.99) | 0.68 ± 0.19 | 0.45 ± 0.22 | 0.61 (0.16–2.31) | ||
| ABCB1—rs2032582 | |||||||||
| CC | 23 | 0.73 ± 0.11 | 0.65 ± 0.12 | 1 (reference) | 0.253 | 0.45 ± 0.11 | 0.45 ± 0.11 | 1 (reference) | 0.59 |
| CT/CA | 21 | 0.83 ± 0.09 | 0.64 ± 0.14 | 1.06 (0.35–3.17) | 0.49 ± 0.12 | 0.49 ± 0.12 | 0.9 (0.39–2.09) | ||
| TT/TA | 5 | 1.00 ± 0.00 | 1.00 ± 1.00 | 0 | 0.80 ± 0.18 | 0.53 ± 0.25 | 0.462 (0.1–2.08) | ||
| ABCB1—rs1128503 | |||||||||
| GG | 22 | 0.70 ± 0.12 | 0.59 ± 0.14 | 1 (reference) | 0.316 | 0.42 ± 0.11 | 0.42 ± 0.11 | 1 (reference) | 0.552 |
| AG | 22 | 0.85 ± 0.08 | 0.70 ± 0.12 | 0.62 (0.2–1.89) | 0.53 ± 0.12 | 0.53 ± 0.12 | 0.71 (0.3–1.65) | ||
| AA | 6 | 1.00 ± 0.00 | 0.80 ± 0.18 | 0.23 (0.03–1.93) | 0.67 ± 0.19 | 0.44 ± 0.22 | 0.54 (0.15–1.95) | ||
| ABCC2—rs3740066 | |||||||||
| CC | 15 | 0.87 ± 0.09 | 0.75 ± 0.13 | 1 (reference) | 0.049 | 0.60 ± 0.13 | 0.49 ± 0.14 | 1 (reference) | 0.471 |
| CT | 23 | 0.88 ± 0.08 | 0.80 ± 0.11 | 1.19 (0.29–5.02) | 0.45 ± 0.12 | 0.45 ± 0.12 | 1.01 (0.4–2.53) | ||
| TT | 9 | 0.70 ± 0.18 | 0.25 ± 0.20 | 4.97 (1.01–24.4) | 0.39 ± 0.17 | NR | 1.89 (0.59–5.96) | ||
| CC/CT | 38 | 0.88 ± 0.06 | 0.78 ± 0.08 | 4.4 (1.21–16.31) | 0.014 | 0.52 ± 0.09 | 0.46 ± 0.09 | 1.86 (0.68 (5.13) | 0.220 |
| ABCC2—rs2273697 | |||||||||
| GG | 33 | 0.80 ± 0.08 | 0.56 ± 0.12 | 1 (reference) | 0.092 | 0.47 ± 0.10 | 0.47 ± 0.10 | 1 (reference) | 0.125 |
| AG | 14 | 0.91 ± 0.08 | 0.91 ± 0.08 | 0.33 (0.07–1.53) | 0.56 ± 0.13 | 0.44 ± 0.15 | 0.91 (0.37–2.25) | ||
| AA | 3 | 0.33 ± 0.27 | 0.33 ± 0.27 | 2.5 (0.54–11.67) | 0.33 ± 0.27 | 0.33 ± 0.27 | 3.24 (0.9–11.58) | ||
| GG/GA | 47 | 0.84 ± 0.06 | 0.68 ± 0.09 | 3.36 (0.74–15.2) | 0.095 | 0.50 ± 0.08 | 0.45 ± 0.09 | 3.35 (0.97–11.51) | 0.042 |
| NQO1—rs1800566 | |||||||||
| GG | 31 | 0.81 ± 0.08 | 0.59 ± 0.11 | 1 (reference) | 0.486 | 0.50 ± 0.10 | 0.42 ± 0.11 | 1 (reference) | 0.325 |
| AG | 15 | 0.76 ± 0.12 | 0.76 ± 0.12 | 0.79 (0.25–2.55) | 0.56 ± 0.14 | 0.56 ± 0.14 | 0.769 (0.3–1.96) | ||
| AA | 4 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0 | NR | NR | 2.17 (0.61–7.69) | ||
| CBR3—rs1056892 | |||||||||
| GG | 28 | 0.82 ± 0.08 | 0.67 ± 0.12 | 1 (reference) | 0.33 | 0.41 ± 0.11 | 0.30 ± 0.12 | 1 (reference) | 0.484 |
| AG | 16 | 0.72 ± 0.12 | 0.62 ± 0.14 | 1.8 (0.62–5.52) | 0.55 ± 0.13 | 0.55 ± 0.13 | 0.87 (0.36–2.06) | ||
| AA | 6 | 1.00 ± 0.00 | 0.75 ± 0.22 | 0.5 (0.06–4.42) | 0.67 ± 0.19 | 0.67 ± 0.19 | 0.41 (0.09–1.82) | ||
| SLC22A16—rs6907567 | |||||||||
| AA | 28 | 0.76 ± 0.09 | 0.60 ± 0.11 | 1 (reference) | 0.49 | 0.48 ± 0.10 | 0.42 ± 0.11 | 1 (reference) | 0.279 |
| AG | 16 | 0.94 ± 0.06 | 0.94 ± 0.06 | 0.44 (0.09–2) | 0.61 ± 0.13 | 0.61 ± 0.13 | 0.71 (0.37–2.26) | ||
| GG | 6 | 0.75 ± 0.22 | 0.38 ± 0.29 | 1.22 (0.27–5.67) | NR | NR | 2.29 (0.73–7.22) | ||
| SLC22A16—rs12210538 | |||||||||
| AA | 38 | 0.81 ± 0.07 | 0.61 ± 0.10 | 1 (reference) | 0.163 | 0.48 ± 0.08 | 0.48 ± 0.08 | 1 (reference) | 0.626 |
| AG | 8 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0 | 0.67 ± 0.20 | 0.33 ± 0.26 | 0.58 (0.17–1.96) | ||
| GG | 4 | 0.50 ± 0.25 | 0.50 ± 0.25 | 1.65 (0.37–7.42) | 0.36 ± 0.28 | 0.36 ± 0.28 | 1.23 (0.28–5.31) | ||
| IFOSFAMIDE | |||||||||
| ALDH1A1—rs3764435 | |||||||||
| AA | 22 | 0.69 ± 0.11 | 0.38 ± 0.09 | 1 (reference) | 0.062 | 0.25 ± 0.10 | 0.25 ± 0.10 | 0.085 | |
| AC | 29 | 0.76 ± 0.09 | 0.65 ± 0.10 | 0.56 (0.23–1.33) | 0.50 ± 0.10 | 0.50 ± 0.10 | |||
| CC | 17 | 0.88 ± 0.08 | 0.80 ± 0.11 | 0.24 (0.06–0.88) | 0.63 ± 0.12 | 0.53 ± 0.12 | |||
| AC/CC | 46 | 0.81 ± 0.06 | 0.71 ± 0.08 | 2.29 (1.02–5.17) | 0.038 | 0.55 ± 0.08 | 0.51 ± 0.08 | 2.04 (1.04–3.99) | 0.034 |
| ALDH1A1—rs168351 | |||||||||
| AA | 53 | 0.81 ± 0.06 | 0.63 ± 0.08 | 1 (reference) | 0.015 | 0.46 ± 0.07 | 0.43 ± 0.07 | 1 (reference) | 0.306 |
| AG | 14 | 0.67 ± 0.14 | 0.56 ± 0.15 | 1.64 (0.65–4.17) | 0.46 ± 0.16 | 0.46 ± 0.16 | 1.14 (0.49–2.61) | ||
| GG | 1 | NR | NR | 11.8 (1.4–99.7) | NR | NR | 4.31 (0.56–33.01) | ||
| AG/GG | 15 | 0.62 ± 0.14 | 0.52 ± 0.15 | 1.86 (0.77–4.5) | 0.16 | 0.42 ± 0.15 | 0.42 ± 0.15 | 1.25 (0.57–2.75) | 0.575 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virgili Manrique, A.C.; Salazar, J.; Arranz, M.J.; Bagué, S.; Orellana, R.; López-Pousa, A.; Cerdà, P.; Gracia, I.; Majercakova, K.; Peiró, A.; et al. Pharmacogenetic Profiling in High-Risk Soft Tissue Sarcomas Treated with Neoadjuvant Chemotherapy. J. Pers. Med. 2022, 12, 618. https://doi.org/10.3390/jpm12040618
Virgili Manrique AC, Salazar J, Arranz MJ, Bagué S, Orellana R, López-Pousa A, Cerdà P, Gracia I, Majercakova K, Peiró A, et al. Pharmacogenetic Profiling in High-Risk Soft Tissue Sarcomas Treated with Neoadjuvant Chemotherapy. Journal of Personalized Medicine. 2022; 12(4):618. https://doi.org/10.3390/jpm12040618
Chicago/Turabian StyleVirgili Manrique, Anna C., Juliana Salazar, María Jesús Arranz, Silvia Bagué, Ruth Orellana, Antonio López-Pousa, Paula Cerdà, Isidre Gracia, Katarina Majercakova, Ana Peiró, and et al. 2022. "Pharmacogenetic Profiling in High-Risk Soft Tissue Sarcomas Treated with Neoadjuvant Chemotherapy" Journal of Personalized Medicine 12, no. 4: 618. https://doi.org/10.3390/jpm12040618
APA StyleVirgili Manrique, A. C., Salazar, J., Arranz, M. J., Bagué, S., Orellana, R., López-Pousa, A., Cerdà, P., Gracia, I., Majercakova, K., Peiró, A., Trullols, L., Fernández, M., Valverde, S., Quintana, M. J., Bell, O., Artigas-Baleri, A., & Sebio, A. (2022). Pharmacogenetic Profiling in High-Risk Soft Tissue Sarcomas Treated with Neoadjuvant Chemotherapy. Journal of Personalized Medicine, 12(4), 618. https://doi.org/10.3390/jpm12040618