Evaluating the Impact of Post-Emergence Weed Control in Honeybee Colonies Located in Different Agricultural Surroundings
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
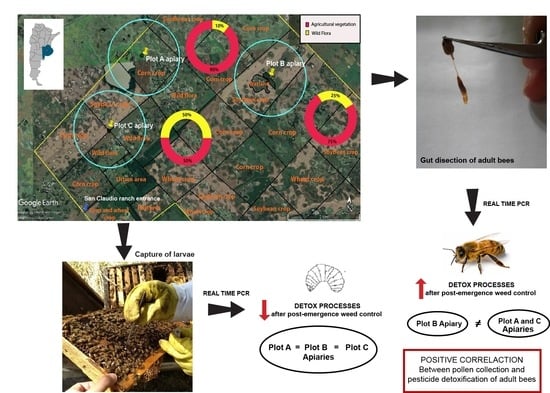
2.1. Study Site and Animals
2.2. Colony Activity
2.3. Sample Collection
2.4. Gene Expression Analysis
2.5. Statistics
3. Results
3.1. Colony Activity
3.2. Relative Gene Expression
3.3. Correlation between Variables Studied
3.4. Principal Component Analysis (PCA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aizen, M.A.; Garibaldi, L.A.; Dondo, M. Expansión de la soja y diversidad de la agricultura argentina. Ecol. Austral. 2009, 19, 45–54. [Google Scholar]
- Aizen, M.A.; Harder, L.D. The global stock of domesticated honey bees is growing lower than agricultural demand for pollination. Curr. Biol. 2009, 19, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Breeze, T.D.; Vaissière, B.E.; Bommarco, R.; Petanidou, T.; Seraphides, N.; Kozák, L.; Scheper, J.; Biesmeijer, J.C.; Kleijn, D.; Gyldenkaerne, S.; et al. Agricultural policies exacerbate honeybee pollination service supply-demand mismatches across Europe. PLoS ONE 2014, 9, e82996. [Google Scholar] [CrossRef] [Green Version]
- Potts, S.G.; Roberts, S.P.M.; Dean, R.; Marris, G.; Brown, M.A.; Jones, R.; Neumann, P.; Settele, J. Declines of managed honey bees and beekeepers in Europe. J. Apic. Res. 2010, 49, 15–22. [Google Scholar] [CrossRef]
- González-Varo, J.P.; Biesmeijer, J.C.; Bommarco, R.; Potts, S.G.; Schweiger, O.; Smith, H.G.; Steffan-Dewenter, I.; Szentgyörgyi, H.; Woyciechowski, M.; Vilà, M. Combined effects of global change pressures on animal-mediated pollination. Trends Ecol. Evol. 2013, 28, 524–530. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Bayo, F.; Goka, K. Pesticide residues and bees—A risk assessment. PLoS ONE 2014, 9, e94482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodschneider, R.; Crailsheim, K. Nutrition and health in honey bees. Apidologie 2010, 41, 278–294. [Google Scholar] [CrossRef]
- Ucar, T.; Hall, F.R. Windbreaks as a pesticide drift mitigation strategy: A review. Pest. Manag. Sci. 2001, 57, 663–675. [Google Scholar] [CrossRef]
- IPBES. The Assessment Report of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services on Pollinators, Pollination and Food Production; Potts, S.G., Imperatriz-Fonseca, V.L., Ngo, H.T., Eds.; Secretariat of the IPBES: Bonn, Germany, 2016; 552p. [Google Scholar] [CrossRef]
- Bordier, C.; Klein, S.; Le Conte, Y.; Barron, A.; Alaux, C. Stress decreases pollen foraging performance in honeybees. J. Exp. Biol. 2018, 221, jeb171470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prado, A.; Pioz, M.; Vidau, C.; Requier, F.; Jury, M.; Crauser, D.; Brunet, J.-L.; Le Conte, Y.; Alaux, C. Exposure to pollen-bound pesticide mixtures induces longer-lived but less efficient honey bees. Sci. Total Environ. 2019, 650, 1250–1260. [Google Scholar] [CrossRef]
- Smart, M.; Pettis, J.; Rice, N.; Browning, Z.; Spivak, M. Linking Measures of Colony and Individual Honey Bee Health to Survival among Apiaries Exposed to Var-ying Agricultural Land Use. PLoS ONE 2016, 11, e0152685. [Google Scholar] [CrossRef] [Green Version]
- Valverde, B.E. Weed management in Latin America. Pestic. Outlook 2002, 13, 79–81. [Google Scholar] [CrossRef]
- Aapresid. Evolución y retos de la siembra directa en Argentina. 2018. Available online: https://www.aapresid.org.ar/wp-content/uploads/2018/03/.
- CASAFE (Cámara de Sanidad Agropecuaria y Fertilizantes). 2014. Available online: www.casafe.org (accessed on 1 May 2020).
- Duke, S.O.; Powles, S.B. Glyphosate: A once in a century herbicide. Pest. Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Song, Y. Insight into the mode of action of 2,4-dichlorophenoxyacetic acid (2,4-D) as an herbicide. J. Integr. Plant Biol. 2014, 56, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Cheremisinoff, N.P.; Rosenfeld, P.E. Handbook of Pollution Prevention and Cleaner Production: Best Practices in the Agrochemical Industry; William Andrew Publishing: Cham, Switzerland, 2007; pp. 215–231. [Google Scholar]
- Cid, R. Aplicación eficiente de fitosanitarios. In Buenas Prácticas en la Aplicación de Fitosanitarios; Instituto Nacional de Tecnología Agropecuaria (INTA): Buenos Aires, Argentina, 2014. [Google Scholar]
- Farina, W.M.; Balbuena, M.S.; Herbert, L.T.; Mengoni Goñalons, C.; Vázquez, D.E. Effects of the herbicide glyphosate on honey bee sensory and cognitive abilities: Individual impairments with implications for the hive. Insects 2019, 10, 354. [Google Scholar] [CrossRef] [Green Version]
- Devillers, J.; Pham-Delègue, H. Honey Bees: Estimating the Environmental Impact of Chemicals; Taylor & Francis: London, UK, 2002. [Google Scholar]
- Fagúndez, G.A.; Blettler, D.C.; Krumrick, C.G.; Bertos, M.A.; Trujillo, C.G. Do agrochemicals used during soybean flowering affect the visits of Apis mellifera L.? Span. J. Agric. Res. 2016, 14, e0301. [Google Scholar] [CrossRef] [Green Version]
- Liao, L.; Wu, W.; Berenbaum, M.R. Behavioral responses of honey bee (Apis mellifera) to natural and synthetic xenobiotics in food. Sci. Rep. 2017, 7, 15924. [Google Scholar] [CrossRef]
- Herbert, L.T.; Vazquez, D.E.; Arenas, A.; Farina, W.M. Effects of field-realistic doses of glyphosate on honeybee appetitive behaviour. J. Exp. Biol. 2014, 217, 3457–3464. [Google Scholar] [CrossRef] [Green Version]
- Balbuena, M.S.; Tison, L.; Hahn, M.-L.; Greggers, U.; Menzel, R.; Farina, W.M. Effects of sublethal doses of glyphosate on honeybee navigation. J. Exp. Biol. 2015, 218, 2799–2805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mengoni Goñalons, C.M.; Farina, W.M. Impaired associative learning after chronic exposure to pesticides in young adult honey bees. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef] [Green Version]
- Seeley, T.D. The Wisdom of the Hive: The Social Physiology of Honey Bee Colonies; Harvard University Press: Cambridge, UK, 1995. [Google Scholar]
- Rortais, A.; Arnold, G.; Halm, M.-P.; Touffet-Briens, F. Modes of honeybees exposure to systemic insecticides: Estimated amounts of contaminated pollen and nec-tar consumed by different categories of bees. Apidologie 2005, 36, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Tosi, S.; Costa, C.; Vesco, U.; Quaglia, G.; Guido, G. A 3-year survey of Italian honey bee collected pollen reveals widespread contamination by agricultural pesticides. Sci. Total Environ. 2018, 615, 208–218. [Google Scholar] [CrossRef]
- Rubio, F.; Guo, E.; Kamp, L. Survey of glyphosate residues in honey, corn and soy products. J. Environ. Anal. Toxicol. 2014, 5, 249. [Google Scholar] [CrossRef]
- Chamkasem, N.; Vargo, J.D. Development and independent laboratory validation of an analytical method for the direct determination of glyphosate, glufosinate, and aminomethylphosphonic acid in honey by liquid chromatography/tandem mass spectrometry. J. Regul. Sci. 2017, 5, 1–9. [Google Scholar] [CrossRef]
- Berg, C.J.; King, H.P.; Delenstarr, G.; Kumar, R.; Rubio, F.; Glaze, T. Glyphosate residue concentra-tions in honey attributed through geospatial analysis to proximity of large-scale agriculture and transfer off-site by bees. PLoS ONE 2018, 13, e0198876. [Google Scholar] [CrossRef]
- Bommuraj, V.; Chen, Y.; Klein, H.; Sperling, R.; Barel, S.; Shimshoni, J.A. Pesticide and trace ele-ment residues in honey and beeswax combs from Israel in association with human risk assessment and honey adulteration. Food Chem. 2019, 299, e125123. [Google Scholar] [CrossRef]
- Mullin, C.A.; Frazier, M.; Frazier, J.L.; Ashcraft, S.; Simonds, R.; van Engelsdorp, D.; Pettis, J.S. High levels of miticides and agrochemicals in North American apiaries: Implications for honey bee health. PLoS ONE 2010, 5, e9754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medrzycki, P.; Giffard, H.; Aupinel, P.; Belzunces, L.P.; Chauzat, M.-P.; Claßen, C.; Colin, M.E.; Dupont, T.; Girolami, V.; Johnson, R.; et al. Standard methods for toxicology research in Apis mellifera. J. Apic. Res. 2008, 52, 1–60. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, D.E.; Latorre-Estivalis, J.M.; Ons, S.; Farina, W.M. Chronic exposure to glyphosate induces transcriptional changes in honey bee larva: A toxicogenomic study. Environ. Pollut. 2020, 261, 114148. [Google Scholar] [CrossRef]
- Boncristiani, H.; Underwood, R.; Schwarz, R.; Evans, J.D.; Pettis, J.; van Engelsdorp, D. Direct effect of acaricides on pathogen loads and gene expression levels in honey bees Apis mellifera. J. Insect Physiol. 2012, 58, 613–620. [Google Scholar] [CrossRef]
- Johnson, R.M.; Mao, W.; Pollock, H.S.; Niu, G.; Schuler, M.A.; Berenbaum, M.R. Ecologically ap-propriate xenobiotics induce cytochrome P450s in Apis mellifera. PLoS ONE 2012, 7, 31051. [Google Scholar] [CrossRef]
- Mao, W.; Schuler, M.A.; Berenbaum, M.R. CYP9Q-mediated detoxification of acaricides in the honey bee (Apis mellifera). Proc. Natl. Acad. Sci. USA 2011, 108, 12657–12662. [Google Scholar] [CrossRef] [Green Version]
- Schmehl, D.R.; Teal, P.E.; Frazier, J.L.; Grozinger, C.M. Genomic analysis of the interaction between pesticide exposure and nutrition in honey bees (Apis mellifera). J. Insect Physiol. 2014, 71, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, D.E.; Ilina, N.; Pagano, E.A.; Zavala, J.A.; Farina, W.M. Glyphosate affects the larval development of honey bees depending on the susceptibility of colonies. PLoS ONE 2018, 13, e0205074. [Google Scholar] [CrossRef] [Green Version]
- Gregorc, A.; Evans, J.D.; Scharf, M.; Ellis, J.D. Gene expression in honey bee (Apis mellifera) larvae exposed to pesticides and Varroa mites (Varroa destructor). J. Insect Physiol. 2012, 58, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Jumarie, C.; Aras, P.; Boily, M. Mixtures of herbicides and metals affect the redox system of honey bees. Chemosphere 2017, 168, 163–170. [Google Scholar] [CrossRef]
- Boily, M.; Sarrasin, B.; Deblois, C.; Aras, P.; Chagnon, M. Acetylcholinesterase in honey bees (Apis mellifera) exposed to neonicotinoids, atrazine and glyphosate: Laboratory and field experiments. Env. Sci. Pollut. Res. Int. 2013, 20, 5603–5614. [Google Scholar] [CrossRef]
- Helmer, S.H.; Kerbaol, A.; Aras, P.; Jumarie, C.; Boily, M. Effects of realistic doses of atrazine, metolachlor, and glyphosate on lipid peroxidation and diet-derived antioxidants in caged honey bees (Apis mellifera). Environ. Sci. Pollut. Res. 2015, 22, 8010–8021. [Google Scholar] [CrossRef]
- Mao, W.; Schuler, M.A.; Berenbaum, M.R. Honey constituents upregulate detoxification and im-munity genes in the western honey bee Apis mellifera. Proc. Natl. Acad. Sci. USA 2013, 110, 8842–8846. [Google Scholar] [CrossRef] [Green Version]
- Berenbaum, M.R.; Johnson, R.M. Xenobiotic detoxification pathways in honey bees. Curr. Opin. Insect Sci. 2015, 10, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defense mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, S.; Armitage, S.A.O.; Schmid-Hempel, P. Social immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grozinger, C.M.; Zayed, A. Improving bee health through genomics. Nat. Rev. Genet. 2020, 21, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Chiari, W.C.; de Alencar Arnaut de Toledo, V.; Ruvolo-Takasusuki, M.C.C.; Braz de Oliveira, A.J.; Shigue-ro Sakaguti, E. Pollination of soybean (Glycine max L. Merril) by honeybees (Apis mellifera L.). Braz. Arch. Biol. Technol. 2005, 48, 31–36. [Google Scholar] [CrossRef]
- Somerville, D.C. Nutritional Value of Bee Collected Pollens; The Rural Industries Research and Development Corporation: Wagga Wagga, NSW, Australia, 2001. [Google Scholar]
- Fehr, W.R.; Caviness, C.F.; Burmood, D.T.; Pennington, J.S. Stage of Development Descriptions for Soybeans, Glycine max (L.) Merrill. Crop. Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Ritchie, S.W.; Hanway, J.J. How a Corn Plant Develops; Iowa State University of Science and Technology, Cooperative Extension Services: Ames, IA, USA, 1982; Special Report 48; 21p. [Google Scholar]
- Schneiter, A.; Miller, J.F. Description of sunflower growth stages. Crop. Sci. 1981, 21, 901–903. [Google Scholar] [CrossRef]
- Biosystems, A. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. 2014. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_042380.pdf (accessed on 18 January 2021).
- Collins, A.; Williams, V.; Evans, J. Sperm storage and antioxidative enzyme expression in the honey bee, Apis Mellifera. Insect Mol. Biol. 2004, 13, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Grosso, J.P.; Barneto, J.A.; Velarde, R.A.; Pagano, E.A.; Zavala, J.A.; Farina, W.M. An Early Sensi-tive Period Induces Long-Lasting Plasticity in the Honeybee Nervous System. Front. Behav. Neurosci. 2018, 12, 11. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 18 January 2021).
- Pohlert, T. PMCMRplus: Calculate Pairwise Multiple Comparisons of Mean Rank Sums Extended, R package version 1.4.4; 2020. Available online: http://CRAN.R-project.org/package=PMCMRplus (accessed on 18 January 2021).
- Le, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Morton, H.L.; Moffett, J.O.; Macdonald, R.H. Toxicity of Herbicides to Newly Emerged Honey Bees. Environ. Entomol. 1972, 1, 102–104. [Google Scholar] [CrossRef]
- Blot, N.; Veillat, L.; Rouzé, R.; Delatte, H. Glyphosate, but not its metabolite AMPA, alters the honeybee gut microbiota. PLoS ONE 2019, 14, e0215466. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Yan, Z.; Ma, S.; Yang, Y.; Wang, Q.; Hou, C.; Wu, Y.; Liu, Y.; Diao, Q. The herbicide glyphosate negatively affects midgut bacterial communities and survival of honey bee during larvae reared in vitro. J. Agric. Food Chem. 2018, 66, 7786–7793. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef] [Green Version]
- Claudianos, C.; Ranson, H.; Johnson, R.M.; Biswas, S.; Schuler, M.A. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef] [Green Version]
- DeGrandi-Hoffman, G.; Martin, J.H. The size and distribution of the honey bee (Apis mellifera L.) cross-pollinating population on male-sterile sunflowers (Helianthus annuus L.). J. Apic. Res. 1993, 32, 135–142. [Google Scholar] [CrossRef]
- Grüter, C.; Farina, W.M. Nectar distribution and its relation to food quality in honeybee (Apis mellifera) colonies. Insectes Sociaux 2007, 54, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Jablonowski, N.D.; Schäffer, A.; Burauel, P. Still present after all these years: Persistence plus po-tential toxicity raise questions about the use of atrazine. Environ. Sci. Pollut. Res. 2011, 18, 328–331. [Google Scholar] [CrossRef] [Green Version]
- Walters, J. Environmental Fate of 2,4-Dichlorophenoxyacetic Acid; Environmental Monitoring and Pest Management, Department of Pesticide Regulation: Sacramento, CA, USA, 2011. Available online: http://www.cdpr.ca.gov/docs/emon/pubs/fatememo/24-d.pdf (accessed on 18 January 2021).
- Ordaz-Guillén, Y.; Galíndez-Mayer, C.J.; Ruiz-Ordaz, N.; Juárez-Ramírez, C.; Santoyo-Tepole, F.; Ramos-Monroy, O. Evaluating the degradation of the herbicides picloram and 2,4-D in a compartmentalized reactive biobarrier with internal liquid recirculation. Environ. Sci. Pollut. Res. 2014, 21, 8765–8773. [Google Scholar] [CrossRef]
- Bento, C.P.M.; Yang, X.; Gort, G.; Xue, S.; van Dam, R.; Zomer, P.; Mol, H.G.J.; Ritsema, C.J.; Geissen, V. Persistence of glyphosate and aminomethylphosphonic acid in loess soil under different combinations of temperature, soil moisture and light/darkness. Sci. Total Environ. 2016, 572, 301–311. [Google Scholar] [CrossRef]
- Raina-Fulton, R. A review of methods for the analysis of orphan and difficult pesticides: Glyphosate, glufosinate, quaternary ammonium and phenoxy acid herbicides, and dithiocarbamate and phthalimide fungicides. J. AOAC Int. 2014, 97, 965–977. [Google Scholar] [CrossRef]
- Alaux, C.; Dantec, C.; Parrinello, H.; Le Conte, Y. Nutrigenomics in honey bees: Digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genom. 2011, 12, 496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corby-Harris, V.; Jones, B.M.; Walton, A.; Schwan, M.R.; Anderson, K.E. Transcriptional markers of suboptimal nutrition in developing Apis mellifera nurse workers. BMC Genom. 2014, 15, 134. [Google Scholar] [CrossRef] [Green Version]
- Decourtye, A.; Mader, E.; Desneux, N. Landscape enhancement of floral resources for honey bees in agro-ecosystems. Apidologie 2010, 41, 264–277. [Google Scholar] [CrossRef] [Green Version]
- Ricketts, T.H.; Regetz, J.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Bogdanski, A.; Gemmill-Herren, B.; Greenleaf, S.S.; Klein, A.M.; Mayfield, M.M.; et al. Landscape effects on crop pollination services: Are there general patterns? Ecol. Lett. 2008, 11, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Tomé, H.V.V.; Schmehl, D.R.; Wedde, A.E.; Godoy, R.S.M.; Ravaiano, S.V.; Guedes, R.N.; Martins, G.F.; Ellis, J.D. Frequently encountered pesticides can cause multiple disorders in developing worker honey bees. Environ. Pollut. 2020, 256, 113420. [Google Scholar] [CrossRef]
- Keller, I.; Fluri, P.; Imdorf, A. Pollen nutrition and colony development in honey bees. Bee World 2005, 86. [Google Scholar] [CrossRef]






| Primer | Sequence (5′ to 3′) | Product Size (MW) | Tm |
|---|---|---|---|
| Rpl8 | F: CACACGGTGGTGGTAATCAT R: CTCGGATTCTTCCTGTACGA | 114 pb | 59 |
| Abaecin | F: CACACTCGAGGTCTGTAGTATCT R: AATGCTGCGCATATCGTGG | 111 pb | 59 |
| CYP6BD1 | F: CTTCTGTTGCTTTTGGAATTCAAGT R: TGCATGCTGCGAGAAAATGT | 106 pb | 59 |
| CYP9Q3 | F: TTGCAAGCTCCATTCGGACA R: AACGGCCACGAATACGGTTA | 130 pb | 60 |
| CYP6AS2 | F: CGCCAGTAGACATCCCATGA R: CTGACGACATGTGTGATCAGTT | 136 pb | 59 |
| CYP6AS3 | F: GCGCGAACACTCCACCA R: CTCGTCCTCGGTACGATTTTACA | 146 pb | 60 |
| CYP6AS4 | F: TTGCGCTCTCATCTCACTCG R: AAATCGCGACAAATGCGGTT | 125 pb | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macri, I.N.; Vázquez, D.E.; Pagano, E.A.; Zavala, J.A.; Farina, W.M. Evaluating the Impact of Post-Emergence Weed Control in Honeybee Colonies Located in Different Agricultural Surroundings. Insects 2021, 12, 163. https://doi.org/10.3390/insects12020163
Macri IN, Vázquez DE, Pagano EA, Zavala JA, Farina WM. Evaluating the Impact of Post-Emergence Weed Control in Honeybee Colonies Located in Different Agricultural Surroundings. Insects. 2021; 12(2):163. https://doi.org/10.3390/insects12020163
Chicago/Turabian StyleMacri, Ivana N., Diego E. Vázquez, Eduardo A. Pagano, Jorge A. Zavala, and Walter M. Farina. 2021. "Evaluating the Impact of Post-Emergence Weed Control in Honeybee Colonies Located in Different Agricultural Surroundings" Insects 12, no. 2: 163. https://doi.org/10.3390/insects12020163
APA StyleMacri, I. N., Vázquez, D. E., Pagano, E. A., Zavala, J. A., & Farina, W. M. (2021). Evaluating the Impact of Post-Emergence Weed Control in Honeybee Colonies Located in Different Agricultural Surroundings. Insects, 12(2), 163. https://doi.org/10.3390/insects12020163