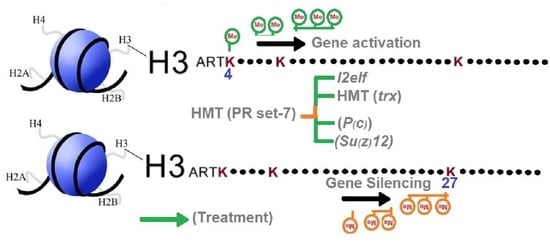
Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Honeybee Sample Preparation
2.2. Cross-Linking, Quenching and Chromatin Isolation
2.3. Chromatin Immunoprecipitation (ChIP)
2.4. Primer Design
2.5. Quantitative Polymerase Chain Reaction (qPCR)
2.6. Statistical Analysis
3. Results
3.1. Apis mellifera (l(2)efl)
3.2. Apis mellifera histone-lysine N-methyltransferase (HMT) ((trx) and PR-set7)
3.3. Apis mellifera Polycomb (Pc) and (Su(z)12)
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- EFSA (European Food Safety Authority). Towards an integrated environmental risk assessment of multiple stressors on bees: Review of research projects in Europe, knowledge gaps and recommendations. EFSA J. 2014, 12, 3594. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 125–957. [Google Scholar] [CrossRef] [PubMed]
- Havard, T.; Laurent, M.; Chauzat, M.P. Impact of Stressors on Honey Bees (Apis mellifera; Hymenoptera: Apidae): Some Guidance for Research Emerge from a Meta-Analysis. Diversity 2020, 12, 7. [Google Scholar] [CrossRef]
- Alattal, Y.; Alghamdi, A. Impact of temperature extremes on survival of indigenous and exotic honey bee subspecies, Apis mellifera, under desert and semiarid climates. Bull. Insectol. 2015, 68, 219–222. [Google Scholar]
- Li, X.; Ma, W.; Jiang, Y. Honeybees (Hymenoptera: Apidae) Adapt to the Shock of High Temperature and High Humidity Through Changes in Sugars and Polyols and Free Amino Acids. J. Insect Sci. 2023, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Southwick, E.E. The honey bee cluster as a homeothermic superorganism. Comp. Biochem. Physiol. Part A Physiol. 1983, 75, 641–645. [Google Scholar] [CrossRef]
- Severson, D.W.; Erickson, E.H.; Williamson, J.L.; Aiken, J.M. Heat stress induced enhancement of heat shock protein gene activity in the honey bee (Apis mellifera). Experientia 1990, 46, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, F.; Heller, H.C. Colonial thermoregulation in honey bees (Apis mellifera). J. Comp. Physiol. 1982, 148, 65–76. [Google Scholar] [CrossRef]
- Bordier, C.; Dechatre, H.; Suchail, S.; Peruzzi, M.; Soubeyrand, S.; Pioz, M.; Pélissier, M.; Crauser, D.; Conte, Y.L.; Alaux, C. Le Colony adaptive response to simulated heat waves and consequences at the individual level in honeybees (Apis mellifera). Sci. Rep. 2017, 7, 3760. [Google Scholar] [CrossRef]
- Ruttner, F. Biogeography and Taxonomy of Honeybees, 1st ed.; Springer: Berlin, Germany, 1988; p. 284. ISBN 978-3-64272651-4. [Google Scholar] [CrossRef]
- Alqarni, A.S.; Ali, H.; Iqbal, J.; Owayss, A.A.; Smith, B.H. Expression of heat shock proteins in adult honey bee (Apis mellifera L.) workers under hot-arid subtropical ecosystems. Saudi J. Biol. Sci. 2019, 26, 1372–1376. [Google Scholar] [CrossRef]
- Ilyasov, R.A.; Lee, M.L.; Yunusbaev, U.B.; Nikolenko, A.G.; Kwon, H.-W. Estimation of C-derived introgression into A. m. mellifera colonies in the Russian Urals using microsatellite genotyping. Genes Genom. 2020, 42, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Ilyasov, R.A.; Lee, M.; Takahashi, J.; Kwon, H.W.; Nikolenko, A.G. A revision of subspecies structure of western honey bee Apis mellifera. Saudi J. Biol. Sci. 2020, 27, 3615–3621. [Google Scholar] [CrossRef] [PubMed]
- Alqarni, A.S.; Hannan, M.A.; Owayss, A.A.; Engel, M.S. The indigenous honeybees of Saudi Arabia (Hymenoptera, Apidae, Apis mellifera jemenitica Ruttner): Their natural history and role in beekeeping. ZooKeys 2011, 134, 83–98. [Google Scholar] [CrossRef]
- POMEP. Climatic Data for Saudi Arabia: Presidency of Metrology and Environmental Protection; Ministry of Defense and Aviation: Riyadh, Saudi Arabia, 2020. [Google Scholar]
- Ali, M. Comparative study for evaluating two honey bee races, Apis mellifera jementica (indigenous race) and Apis mellifera carnica (Carniolan race) in brood production, population development and foraging activity under the environmental conditions of the central region of the Kingdom of Saudi Arabia. Ann. Agric. Sci. 2011, 56, 127–134. [Google Scholar] [CrossRef]
- Ali, H.; Alqarni, A.S.; Owayss, A.A.; Hassan, A.M.; Smith, B.S. Osmotic concentration in three races of honey bee, Apis mellifera L. under environmental conditions of arid zone. Saudi J. Biol. Sci. 2017, 24, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Stabentheiner, A.; Kovac, H.; Mandl, M.; Kaefar, H. Coping with the cold and fighting the heat: Thermal homeostasis of a superorganism, the honey bee colony. J. Comp. Physiol. A 2021, 207, 337–351. [Google Scholar] [CrossRef]
- Alghamdi, A.A.; Alattal, Y.Z. Expression Levels of Heat-Shock Proteins in Apis mellifera jemenitica and Apis mellifera carnica Foragers in the Desert Climate of Saudi Arabia. Insects 2023, 14, 432. [Google Scholar] [CrossRef]
- Awad, A.M.; Owayss, A.A.; Alqarni, A.S. Performance of two honey bee subspecies during harsh weather and Acacia gerrardii nectar-rich flow. Sci. Agric. 2017, 74, 474–480. [Google Scholar] [CrossRef]
- Dogantzis, K.A.; Tiwari, T.; Conflitti, I.M.; Dey, A.; Patch, H.M.; Muli, E.M.; Garnery, L.; Whitfield, C.W.; Stolle, E.; Alqarni, A.S. Thrice out of Asia and the adaptive radiation of the western honey bee. Sci. Adv. 2021, 7, eabj2151. [Google Scholar] [CrossRef]
- Elekonich, M.M. Extreme thermotolerance and behavioral induction of 70-kDa heat shock proteins and their encoding genes in honey bees. Cell Stress Chaperon 2009, 14, 219–226. [Google Scholar] [CrossRef]
- Zhao, L.; Jones, W.A. Expression of heat shock protein genes in insect stress responses. Invert. Surviv. J. 2012, 9, 93–101. [Google Scholar]
- Zhao, H.; Li, G.; Guo, D.; Li, H.; Liu, Q.; Xu, B.; Guo, X. Response mechanisms to heat stress in bees. Apidologie 2021, 52, 388–399. [Google Scholar] [CrossRef]
- Huang, C.; Xu, M.; Zhu, B. Epigenetic inheritance mediated by histone lysine methylation: Maintaining transcriptional states without the precise restoration of marks? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20110332. [Google Scholar] [CrossRef] [PubMed]
- Binda, O. On your histone mark, SET, methylate! Epigenetica 2013, 8, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.J.; Kucharski, R.; Maleszka, R.; Hurd, P.J. Extensive histone post-translational modification in honey bees. Insect. Biochem. Mol. Biol. 2013, 43, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Alattal, Y.Z.; Alghamdi, A.A. Linking Histone Methylation States and hsp Transcriptional Regulation in Thermo-Tolerant and Thermo-Susceptible A. mellifera L. Subspecies in Response to Heat Stress. Insects 2023, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Bloch, G.; Grozinger, C.M. Social molecular pathways and the evolution of bee societies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 27, 2155–2170. [Google Scholar] [CrossRef]
- Caparros, M.L.; Allis, C.D.; Jenuwein, T.; Reinberg, D. Epigenetics, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015; p. 984. ISBN 139781936113590. [Google Scholar]
- Viré, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef]
- Ross, P.J.; Ragina, N.P.; Rodriguez, R.M.; Iager, A.E.; Siripattarapravat, K.; Lopez-Corrales, N.; Cibelli, J.B. Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction 2008, 136, 777–785. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 2007, 128, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.H.; Chi, A.S.; et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008, 4, e1000242. [Google Scholar] [CrossRef] [PubMed]
- Runtuwene, L.R.; Kawashima, S.; Pijoh, V.D.; Tuda, J.S.; Hayashida, K.; Yamagishi, J.; Sugimoto, C.; Nishiyama, S.; Sasaki, M.; Orba, Y.; et al. The Lethal(2)-Essential-for-Life [L(2)EFL] Gene Family Modulates Dengue Virus Infection in Aedes aegypti. Int. J. Mol. Sci. 2020, 21, 7520. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep. 2017, 7, 6448. [Google Scholar] [CrossRef] [PubMed]
- Kurzik-Dumke, U.; Lohmann, E. Sequence of the new Drosophila melanogaster small heat-shock-related gene, lethal (2) essential for life [l(2)efl], at locus 59F4, 5. Gene 1995, 154, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, X.; Lu, M.; Du, Y. Characterization of genes encoding small heat shock proteins from Bemisia tabaci and expression under thermal stress. PeerJ 2019, 7, e6992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Xiao, Z.J.; Zeng, B.P.; Li, K.; Tang, Y.L. Insect behavior and physiological adaptation mechanisms under starvation stress. Front. Physiol. 2019, 10, 163. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, L.; Jiang, X. Starvation influences allatotropin gene expression and juvenile hormone titer in the female adult oriental armyworm, Mythimna separata. Arch. Insect Biochem. Physiol. 2008, 68, 63–70. [Google Scholar] [CrossRef]
- Qin, M.; Wang, H.; Liu, Z.; Wang, Y.; Zhang, W.; Xu, B. Changes in cold tolerance during the overwintering period in Apis mellifera ligustica. J. Apicul. Res. 2019, 58, 702–709. [Google Scholar] [CrossRef]
- McAfee, A.; Chapman, A.; Higo, H.; Underwood, R.; Milone, J.; Foster, L.J.; Pettis, J.S. Vulnerability of honey bee queens to heat-induced loss of fertility. Nat. Sustain. 2020, 3, 367–376. [Google Scholar] [CrossRef]
- Yi, J.; Liu, J.; Li, D.; Sun, D.; Li, J.; An, Y.; Wu, H. Transcriptome responses to heat and cold stress in prepupae of Trichogramma chilonis. Ecol. Evol. 2021, 11, 4816–4825. [Google Scholar] [CrossRef]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Strauch, A.; Haslbeck, M. The function of small heat-shock proteins and their implication in proteostasis. Essays Biochem. 2016, 60, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, L.R. Genome Architecture and Phenotypic Plasticity: Is the Lethal (2) Essential for Life Cluster Epigenetically Regulated during Ovary Activation in the Honeybee, Apis mellifera? Master’s Thesis, University of Otago, Dunedin, New Zealand, 2013. [Google Scholar]
- Zhang, Y.; Li, Z.; He, X.; Wang, Z.; Zeng, Z. H3K4me1 Modification Functions in Caste Differentiation in Honey Bees. Int. J. Mol. Sci. 2023, 24, 6217. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.R.; Bach, D.M.; Rondeau, N.C.; Sam, J.; Lovinger, N.L.; Lopatkin, A.J.; Snow, J.W. Honey bee sHSP are responsive to diverse proteostatic stresses and potentially promising biomarkers of honey bee stress. Sci Rep. 2021, 11, 22087. [Google Scholar] [CrossRef] [PubMed]
- Ruttner, H. Technische Empfehlungen zur Leistungsprüfung von Bienenvölkern. In Proceedings of the Paarungskontrolle und Selektion bei der Honigbiene: Internationales Symposium, Lunz am See, Austria, 31 July–5 August 1972. [Google Scholar]
- Milne, T.A.; Zhao, K.; Hess, J.L. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol. Biol. 2009, 538, 409–423. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 1165–1188. [Google Scholar] [CrossRef]
- Xu, X.; Sarbeng, E.B.; Vorvis, C.; Kumar, D.P.; Zhou, L.; Liu, Q. Unique peptide substrate binding properties of 110-kDa heatshock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 2012, 287, 5661–5672. [Google Scholar] [CrossRef]
- Muller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Malatesta, M.; Jung, H.R.; Walfridsson, J.; Willer, A.; Olsson, L.; Helin, K. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010, 38, 4958–4969. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Tang, T.; Song, Q.; Wang, Z.; He, K.; Liu, X.; Song, J.; Wang, L.; Yang, Y.; Feng, C. Transcription Analysis of the Stress and Immune Response Genes to Temperature Stress in Ostrinia furnacalis. Front. Physiol. 2019, 10, 1289. [Google Scholar] [CrossRef]



| Gene ID/Gene Name | Location (LG2)/Length | Primers |
|---|---|---|
| (l(2)efl) (A. mellifera) ID:724405 | (LG2):4831304..4832577 /1274nt | F-TGCGACATCGATCAAGCGTCC R-TTGCGCATCGCACGGTTTCC |
| (l(2)efl) (A. mellifera) ID:724488 | (LG2):4837466..4838343 /878nt | F-ACCTTGGGGTGAACTTCTGCG R-TCCCCTCGACGACAACACAC |
| (l(2)efl) (A. mellifera) ID:724274 | (LG2):4823146..4824181 /1063nt | F-TCACCGAGCCGATTGGAGTTATGT R-AACTGCCTCTGTCACCACGAAAC |
| A. mellifera HLMT (trx) ID: 408716 | (LG2):4633633..4650053, complement | F-TGCAGCTAGATTCATTAATCATTCATGR-CATGGAATCTTGATATCCTCGAAAG |
| A. mellifera Polycomb protein Su(z)12, ID: 409170 | (LG10)11912175..11918682 | F-ATGCTCTGCCCAAGCAACTATTACG R-CGGAACCTCCATCTTGTTACATAAA |
| Apis mellifera HLMT- PR-set7 (PR-set7), ID: 412027 | (LG8) (1813753..1816546) | F-TGGTAAAGGTCGTGGAATAGTAACA R-AGTTTCTGCTGTTGCATCAACGC |
| Apis mellifera Polycomb (Pc), ID:725474. | (LG3) (8393126..8397763, complement) | F-ACAACGTCAAAGCAATGACAAATTA R-AGCTGCTCCAAAATATATGTTCACCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alghamdi, A.A.; Alattal, Y.Z. Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress. Insects 2024, 15, 33. https://doi.org/10.3390/insects15010033
Alghamdi AA, Alattal YZ. Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress. Insects. 2024; 15(1):33. https://doi.org/10.3390/insects15010033
Chicago/Turabian StyleAlghamdi, Ahmad A., and Yehya Z. Alattal. 2024. "Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress" Insects 15, no. 1: 33. https://doi.org/10.3390/insects15010033
APA StyleAlghamdi, A. A., & Alattal, Y. Z. (2024). Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress. Insects, 15(1), 33. https://doi.org/10.3390/insects15010033