Differential Gene Expression in Red Imported Fire Ant (Solenopsis invicta) (Hymenoptera: Formicidae) Larval and Pupal Stages
Abstract
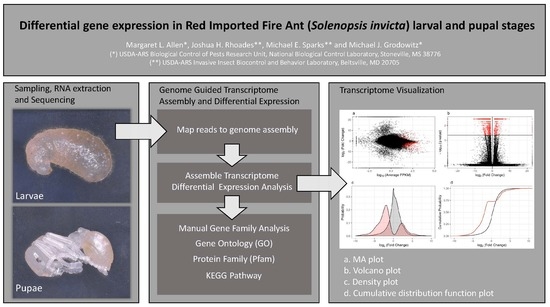
:1. Introduction
2. Materials and Methods
2.1. Colony Origin and Rearing
2.2. Sampling and RNA Extraction
2.3. Sequencing, Transcriptome Assembly and Analysis
3. Results
3.1. Sequencing, Assembly, Differential Expression Analysis and Functional Analysis
3.2. Gene Family Analysis by Functional Category
3.2.1. Digestion, Nutrient Storage and Xenobiotic Detoxification
3.2.2. Muscle, Cuticle and Neuronal Development
3.2.3. Cell Regulation, Hormone Signaling and Fatty Acid Metabolism
3.2.4. Immunity and Caste Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tschinkel, W.R. The Fire Ants; Belknap Press of Harvard University Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Korzukhin, M.D.; Porter, S.D.; Thompson, L.C.; Wiley, S. Modeling temperature-dependent range limits for the fire ant Solenopsis invicta (Hymenoptera: Formicidae) in the United States. Environ. Entomol. 2001, 30, 645–655. [Google Scholar] [CrossRef]
- Allen, C.; Epperson, D.; Garmestani, A. Red imported fire ant impacts on wildlife: A decade of research. Am. Midl. Nat. 2004, 152, 88–103. [Google Scholar] [CrossRef]
- Vinson, S.B. Insect life: Invasion of the red imported fire ant (Hymenoptera: Formicidae). Am. Entomol. 1997, 43, 23–39. [Google Scholar] [CrossRef]
- Taber, S.W. Fire Ants; Texas A&M University Press: College Station, TX, USA, 2000. [Google Scholar]
- Williams, D.F. Chemical baits: Specificity and effects on other ant species. In Fire Ants and Leaf-Cutting Ants: Biology and Management; Lofgren, C.S., Vander Meer, R.K., Eds.; Westview Press: Boulder, CO, USA, 1986; pp. 378–386. [Google Scholar]
- Wurm, Y.; Wang, J.; Riba-Grognuz, O.; Corona, M.; Nygaard, S.; Hunt, B.G.; Ingram, K.K.; Falquet, L.; Nipitwattanaphon, M.; Gotzek, D.; et al. The genome of the fire ant Solenopsis invicta. Proc. Natl. Acad. Sci. USA 2011, 108, 5679–5684. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jemielity, S.; Uva, P.; Wurm, Y.; Graff, J.; Keller, L. An annotated cDNA library and microarray for large-scale gene-expression studies in the ant Solenopsis invicta. Genome Biol. 2007, 8, R9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, S.; Skowronski, D.; Hunt, B.G. Developmental DNA methyltransferase expression in the fire ant Solenopsis invicta. Insect Sci. 2018, 25, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Gadau, J.; Helmkampf, M.; Nygaard, S.; Roux, J.; Simola, D.F.; Smith, C.R.; Suen, G.; Wurm, Y.; Smith, C.D. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 2012, 28, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Roux, J.; Privman, E.; Moretti, S.; Daub, J.T.; Robinson-Rechavi, M.; Keller, L. Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 2014, 31, 1661–1685. [Google Scholar] [CrossRef] [PubMed]
- Simola, D.F.; Wissler, L.; Donahue, G.; Waterhouse, R.M.; Helmkampf, M.; Roux, J.; Nygaard, S.; Glastad, K.M.; Hagen, D.E.; Viljakainen, L.; et al. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 2013, 23, 1235–1247. [Google Scholar] [CrossRef] [Green Version]
- Gotzek, D.; Ross, K.G. Genetic regulation of colony social organization in fire ants: An integrative overview. Q. Rev. Biol. 2007, 82, 201–226. [Google Scholar] [CrossRef]
- Manfredini, F.; Riba-Grognuz, O.; Wurm, Y.; Keller, L.; Shoemaker, D.; Grozinger, C.M. Sociogenomics of cooperation and conflict during colony founding in the fire ant Solenopsis invicta. PLoS Genet. 2013, 9, e1003633. [Google Scholar] [CrossRef] [PubMed]
- Buechel, S.D.; Wurm, Y.; Keller, L. Social chromosome variants differentially affect queen determination and the survival of workers in the fire ant Solenopsis invicta. Mol. Ecol. 2014, 23, 5117–5127. [Google Scholar] [CrossRef] [PubMed]
- Manfredini, F.; Lucas, C.; Nicolas, M.; Keller, L.; Shoemaker, D.; Grozinger, C.M. Molecular and social regulation of worker division of labour in fire ants. Mol. Ecol. 2014, 23, 660–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBoeuf, A.C.; Waridel, P.; Brent, C.S.; Gonçalves, A.N.; Menin, L.; Ortiz, D.; Riba-Grognuz, O.; Koto, A.; Soares, Z.G.; Privman, E. Oral transfer of chemical cues, growth proteins and hormones in social insects. Elife 2016, 5, e20375. [Google Scholar] [CrossRef] [PubMed]
- Wurm, Y.; Wang, J.; Keller, L. Changes in reproductive roles are associated with changes in gene expression in fire ant queens. Mol. Ecol. 2010, 19, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Nipitwattanaphon, M.; Wang, J.; Ross, K.G.; Riba-Grognuz, O.; Wurm, Y.; Khurewathanakul, C.; Keller, L. Effects of ploidy and sex-locus genotype on gene expression patterns in the fire ant Solenopsis invicta. Proc. Biol. Sci. 2014, 281. [Google Scholar] [CrossRef]
- Engsontia, P.; Sangket, U.; Robertson, H.M.; Satasook, C. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res. Notes 2015, 8, 380. [Google Scholar] [CrossRef]
- Zhang, W.; Wanchoo, A.; Ortiz-Urquiza, A.; Xia, Y.; Keyhani, N.O. Tissue, developmental, and caste-specific expression of odorant binding proteins in a eusocial insect, the red imported fire ant, Solenopsis invicta. Sci. Rep. 2016, 6, 35452. [Google Scholar] [CrossRef]
- Wurm, Y.; Uva, P.; Ricci, F.; Wang, J.; Jemielity, S.; Iseli, C.; Falquet, L.; Keller, L. Fourmidable: A database for ant genomics. BMC Genom. 2009, 10, 5. [Google Scholar] [CrossRef]
- Pracana, R.; Levantis, I.; Martinez-Ruiz, C.; Stolle, E.; Priyam, A.; Wurm, Y. Fire ant social chromosomes: Differences in number, sequence and expression of odorant binding proteins. Evol. Lett. 2017, 1, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Nipitwattanaphon, M.; Wang, J.; Dijkstra, M.B.; Keller, L. A simple genetic basis for complex social behaviour mediates widespread gene expression differences. Mol. Ecol. 2013, 22, 3797–3813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, B.G.; Ometto, L.; Keller, L.; Goodisman, M.A.D. Evolution at two levels in fire ants: The relationship between patterns of gene expression and protein sequence evolution. Mol. Biol. Evol. 2013, 30, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Calkins, T.L.; Chen, M.-E.; Arora, A.K.; Hawkings, C.; Tamborindeguy, C.; Pietrantonio, P.V. Brain gene expression analyses in virgin and mated queens of fire ants reveal mating-independent and socially regulated changes. Ecol. Evol. 2018, 8, 4312–4327. [Google Scholar] [CrossRef] [PubMed]
- Dussutour, A.; Simpson, S.J. Communal nutrition in ants. Curr. Biol. 2009, 19, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, S.T.; Blum, M.S.; Travis, J. Proteolytic enzymes from larvae of the fire ant, Solenopsis invicta. Isolation and characterization of four serine endopeptidases. J. Biol. Chem. 1998, 273, 14430–14434. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, S.T.; Kordula, T.; Travis, J. Molecular cloning of Soli E2: An elastase-like serine proteinase from the imported red fire ant (Solenopsis invicta). Insect Biochem. Mol. Biol. 1999, 29, 249–254. [Google Scholar] [CrossRef]
- Howard, D.F.; Tschinkel, W.R. The flow of food in colonies of the fire ant, Solenopsis invicta: A multifactorial study. Physiol. Entomol. 1981, 6, 297–306. [Google Scholar] [CrossRef]
- Sorensen, A.A.; Vinson, S. Quantitative food distribution studies within laboratory colonies of the imported fire ant, Solenopsis invicta Buren. Insect. Soc. 1981, 28, 129–160. [Google Scholar] [CrossRef]
- Petralia, R.; Vinson, S. Feeding in the larvae of the imported fire ant, Solenopsis invicta: Behavior and morphological adaptations. Ann. Entomol. Soc. Am. 1978, 71, 643–648. [Google Scholar] [CrossRef]
- Petralia, R.; Vinson, S. Developmental morphology of larvae and eggs of the imported fire ant, Solenopsis invicta. Ann. Entomol. Soc. Am. 1979, 72, 472–484. [Google Scholar] [CrossRef]
- Petralia, R.S.; Vinson, S. Comparative anatomy of the ventral region of ant larvae, and its relation to feeding behavior. Psyche 1979, 86, 375–394. [Google Scholar] [CrossRef]
- Glancey, B.M.; Vander Meer, R.; Glover, A.; Lofgren, C.; Vinson, S. Filtration of microparticles from liquids ingested by the red imported fire ant Solenopsis invicta Buren. Insect. Soc. 1981, 28, 395–401. [Google Scholar] [CrossRef]
- Hölldobler, B.; Wilson, E.O. The Superorganism: The Beauty, Elegance, and Strangeness of Insect Societies; WW Norton & Company: New York, NY, USA, 2009. [Google Scholar]
- Behmer, S.T. Animal behaviour: Feeding the superorganism. Curr. Biol. 2009, 19, R366–R368. [Google Scholar] [CrossRef] [PubMed]
- Knorr, E.; Fishilevich, E.; Tenbusch, L.; Frey, M.L.; Rangasamy, M.; Billion, A.; Worden, S.E.; Gandra, P.; Arora, K.; Lo, W. Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Sci. Rep. 2018, 8, 2061. [Google Scholar] [CrossRef] [PubMed]
- Gundersen-Rindal, D.E.; Adrianos, S.L.; Allen, M.L.; Becnel, J.J.; Chen, Y.P.; Choi, M.-Y.; Estep, A.; Evans, J.D.; Garczynski, S.F.; Geib, S.M.; et al. Arthropod genomics research in the United States Department of Agriculture, Agricultural Research Service: Applications of RNA interference and CRISPR gene-editing technologies in pest control. Trends Entomol. 2017, 13, 109–137. [Google Scholar]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Vélez, A.M.; Fishilevich, E. The mysteries of insect RNAi: A focus on dsRNA uptake and transport. Pestic. Biochem. Phys. 2018, 151, 25–31. [Google Scholar] [CrossRef]
- Lu, H.L.; Vinson, S.; Pietrantonio, P.V. Oocyte membrane localization of vitellogenin receptor coincides with queen flying age, and receptor silencing by RNAi disrupts egg formation in fire ant virgin queens. FEBS J. 2009, 276, 3110–3123. [Google Scholar] [CrossRef] [Green Version]
- Choi, M.Y.; Vander Meer, R.K.; Coy, M.; Scharf, M.E. Phenotypic impacts of PBAN RNA interference in an ant, Solenopsis invicta, and a moth, Helicoverpa zea. J. Insect Physiol. 2012, 58, 1159–1165. [Google Scholar] [CrossRef]
- Cheng, D.; Lu, Y.; Zeng, L.; Liang, G.; He, X. Si-CSP9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 2015, 5, 9245. [Google Scholar] [CrossRef]
- Vander Meer, R.K.; Choi, M.Y. Control of Insect Pests through RNAi of Pheromone Biosynthesis Activating Neuropeptide Receptor. Google Patents US9000145B2, 2015. [Google Scholar]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Ramaseshadri, P.; Segers, G.; Flannagan, R.; Wiggins, E.; Clinton, W.; Ilagan, O.; McNulty, B.; Clark, T.; Bolognesi, R. Physiological and cellular responses caused by RNAi- mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS ONE 2013, 8, e54270. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A.; Lofgren, C.; Jouvenaz, D.; Stringer, C.; Bishop, P.; Williams, D.; Wojcik, D.; Glancey, B. Techniques for Collecting, Rearing, and Handling Imported Fire Ants; Agricultural Research (Southern Region), Science and Education Administration, USDA: Washington, DC, USA, 1981.
- Chen, J. Advancement on techniques for the separation and maintenance of the red imported fire ant colonies. Insect Sci. 2007, 14, 1–4. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Toprak, U.; Baldwin, D.; Erlandson, M.; Gillott, C.; Hegedus, D.D. Insect intestinal mucins and serine proteases associated with the peritrophic matrix from feeding, starved and moulting Mamestra configurata larvae. Insect Mol. Biol. 2010, 19, 163–175. [Google Scholar] [CrossRef]
- Wheeler, D.E.; Martinez, T. Storage proteins in ants (Hymenoptera: Formicidae). Comp. Biochem. Physiol. Part B Biochem. Mol. Boil. 1995, 112, 15–19. [Google Scholar] [CrossRef]
- Martinez, T.; Wheeler, D. Identification of two storage hexamers in the ant, Camponotus festinatus: Accumulation in adult queenless workers. Insect Biochem. Mol. Biol. 1993, 23, 309–317. [Google Scholar] [CrossRef]
- Martinez, T.; Wheeler, D.E. Storage proteins in adult ants (Camponotus festinatus): Roles in colony founding by queens and in larval rearing by workers. J. Insect Physiol. 1994, 40, 723–729. [Google Scholar] [CrossRef]
- Burmester, T. Evolution and function of the insect hexamerins. Eur. J. Entomol. 1999, 96, 213–226. [Google Scholar]
- Haunerland, N. Insect storage proteins: Gene families and receptors. Insect Biochem. Mol. Biol. 1996, 26, 755–765. [Google Scholar] [CrossRef]
- Eliyahu, D.; Ross, K.G.; Haight, K.L.; Keller, L.; Liebig, J. Venom alkaloid and cuticular hydrocarbon profiles are associated with social organization, queen fertility status, and queen genotype in the fire ant Solenopsis invicta. J. Chem. Ecol. 2011, 37, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Shukla, E.; Thorat, L.J.; Nath, B.B.; Gaikwad, S.M. Insect trehalase: Physiological significance and potential applications. Glycobiology 2015, 25, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Field, L.M.; Devonshire, A.L.; Tyler-Smith, C. Analysis of amplicons containing the esterase genes responsible for insecticide resistance in the peach-potato aphid Myzus persicae (Sulzer). Biochem. J. 1996, 313, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Buček, A.; Brabcová, J.; Vogel, H.; Prchalová, D.; Kindl, J.; Valterová, I.; Pichová, I. Exploring complex pheromone biosynthetic processes in the bumblebee male labial gland by RNA sequencing. Insect Mol. Biol. 2016, 25, 295–314. [Google Scholar] [CrossRef]
- Thweatt, R.; Lumpkin, C.K., Jr.; Goldstein, S. A novel gene encoding a smooth muscle protein is overexpressed in senescent human fibroblasts. Biochem. Biophys. Res. Commun. 1992, 187, 1–7. [Google Scholar] [CrossRef]
- Clark, K.A.; Kadrmas, J.L. Drosophila melanogaster Muscle LIM Protein and α-actinin function together to stabilize muscle cytoarchitecture: A potential role for Mlp84B in actin-crosslinking. Cytoskeleton 2013, 70, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.H. Structural cuticular proteins from arthropods: Annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010, 40, 189–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merzendorfer, H. Insect-Derived Chitinases. In Yellow Biotechnology II: Insect Biotechnology in Plant Protection and Industry; Vilcinskas, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 19–50. [Google Scholar]
- Karasov, W.H.; Douglas, A.E. Comparative digestive physiology. Compr. Physiol. 2013, 3, 741–783. [Google Scholar] [PubMed]
- Charlton-Perkins, M.; Cook, T.A. Building a fly eye: Terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr. Top. Dev. Biol. 2010, 93, 129–173. [Google Scholar]
- Martin, J.H.; Benzer, S.; Rudnicka, M.; Miller, C.A. Calphotin: A Drosophila photoreceptor cell calcium-binding protein. Proc. Nat. Acad. Sci. USA 1993, 90, 1531–1535. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ballinger, D. Mutations in calphotin, the gene encoding a Drosophila photoreceptor cell-specific calcium-binding protein, reveal roles in cellular morphogenesis and survival. Genetics 1994, 138, 413–421. [Google Scholar] [PubMed]
- Nässel, D.R. Neuropeptides in the nervous system of Drosophila and other insects: Multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 2002, 68, 1–84. [Google Scholar] [CrossRef]
- Drapeau, M.D.; Albert, S.; Kucharski, R.; Prusko, C.; Maleszka, R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006, 16, 1385–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buttstedt, A.; Moritz, R.F.; Erler, S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. 2014, 89, 255–269. [Google Scholar] [CrossRef]
- Tschinkel, W.R. Social control of egg-laying rate in queens of the fire ant, Solenopsis invicta. Physiol. Entomol. 1988, 13, 327–350. [Google Scholar] [CrossRef]




| Sample | Total Read Bases (bp) | Total Reads | GC (%) | AT (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| X01wp | 11,021,733,676 | 109,126,076 | 49.37 | 50.63 | 93.77 | 89.38 |
| Y02wp | 11,915,906,472 | 117,979,272 | 49.03 | 50.97 | 92.97 | 88.54 |
| Z03wp | 7,833,054,394 | 77,554,994 | 48.38 | 51.62 | 94.65 | 90.29 |
| A01L4 | 10,596,167,954 | 104,912,554 | 48.95 | 51.05 | 93.83 | 89.57 |
| B02L4 | 10,050,206,394 | 99,506,994 | 46.85 | 53.15 | 93.7 | 89.45 |
| C03L4 | 8,565,121,584 | 84,803,184 | 46.39 | 53.61 | 94.88 | 90.78 |
| Quantity | Symbol | PFamily Annotation | PFamily ID |
|---|---|---|---|
| 1243 | RVT_1 | Reverse transcriptase (RNA-dependent DNA polymerase) | PF00078.25 |
| 849 | Pkinase | Protein kinase domain | PF00069.23 |
| 823 | Pkinase_Tyr | Protein tyrosine kinase | PF07714.15 |
| 676 | rve | Integrase core domain | PF00665.24 |
| 641 | zf-C2H2 | Zinc finger, C2H2 type | PF00096.24 |
| 625 | zf-C2H2_4 | C2H2-type zinc finger | PF13894.4 |
| 512 | RRM_1 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) | PF00076.20 |
| 490 | Ig_3 | Immunoglobulin domain | PF13927.4 |
| 478 | WD40 | WD domain, G-beta repeat | PF00400.30 |
| 473 | zf-H2C2_2 | Zinc-finger double domain | PF13465.4 |
| 472 | I-set | Immunoglobulin I-set domain | PF07679.14 |
| 462 | MFS_1 | Major Facilitator Superfamily | PF07690.14 |
| 455 | Ig_2 | Immunoglobulin domain | PF13895.4 |
| 449 | ig | Immunoglobulin domain | PF00047.23 |
| 426 | 7tm_6 | 7tm Odorant receptor | PF02949.18 |
| 420 | p450 | Cytochrome P450 | PF00067.20 |
| 400 | V-set | Immunoglobulin V-set domain | PF07686.15 |
| 345 | Transposase_1 | Transposase (partial DDE domain) | PF01359.16 |
| 334 | ANAPC4_WD40 | Anaphase-promoting complex subunit 4 WD40 domain | PF12894.5 |
| 327 | DDE_3 | DDE superfamily endonuclease | PF13358.4 |
| Category | Subcategory | Number | >DEG Larvae | >DEG Pupae |
|---|---|---|---|---|
| Metabolic pathways | ||||
| Amino acids | 67 | 67 | 0 | |
| Fats/Lipids | 63 | 47 | 16 | |
| Nucleic acids | 39 | 33 | 6 | |
| Carbon metabolism | 36 | 36 | 0 | |
| Carbohydrates | 35 | 35 | 0 | |
| Detoxification | 33 | 31 | 2 | |
| Vitamins | 19 | 19 | 0 | |
| Amino sugar and nucleotide sugar metabolism | 14 | 14 | 0 | |
| Pyruvate metabolism | 11 | 11 | 0 | |
| Cellular processes | ||||
| Signaling pathways | 42 | 23 | 19 | |
| Membrane trafficking | 16 | 14 | 2 | |
| Genetic information processing | 9 | 5 | 4 | |
| Apoptosis | 6 | 5 | 1 | |
| Organelle Biosystems | ||||
| Peroxisome | 29 | 25 | 4 | |
| Phagosome | 17 | 16 | 1 | |
| Protein processing in endoplasmic reticulum | 15 | 15 | 0 | |
| Lysosome | 14 | 14 | 0 | |
| Ribosome related | 4 | 4 | 0 | |
| Biosynthesis pathways | ||||
| Biosynthesis of amino acids | 23 | 23 | 0 | |
| Fatty acid biosynthesis | 18 | 10 | 8 | |
| Insect hormone biosynthesis | 13 | 12 | 1 | |
| Terpenoid backbone biosynthesis | 10 | 10 | 0 | |
| Degradation pathways | ||||
| Amino acid degradation | 28 | 28 | 0 | |
| Fatty acid degradation | 18 | 18 | 0 | |
| Glycosaminoglycan degradation & other glycan degradation | 7 | 6 | 1 | |
| RNA degradation | 3 | 3 | 0 | |
| Totals | 589 | 524 (89%) | 65 (11%) | |
| Gene ID | Annotation (Predicted) | Larvae | Pupae | Binary Log (Fold Change) | p | q |
|---|---|---|---|---|---|---|
| LOC105199117 | chymotrypsin-1-like | 14,120.80 | 7.70 | −10.84 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105194373 | phospholipase A1-like, partial | 383.05 | 0.22 | −10.77 | 1.05 × 10−3 | 3.38 × 10−2 |
| LOC105199115 | chymotrypsin-1-like | 8648.28 | 5.45 | −10.63 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105193961 | chymotrypsin-1-like | 26,016.40 | 18.53 | −10.46 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105198380 | lipase 3-like, partial | 1058.65 | 0.97 | −10.09 | 2.00 × 10−4 | 8.84 × 10−3 |
| LOC105193957 | chymotrypsin-1-like | 510.44 | 0.81 | −9.30 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105208099 | lipase 3-like | 368.52 | 0.65 | −9.15 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105198043 | alpha-amylase | 2527.41 | 5.70 | −8.79 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105194610 | peritrophin-1-like | 4362.25 | 11.73 | −8.54 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105193995 | chymotrypsin-2-like | 528.99 | 1.64 | −8.33 | 3.00 × 10−4 | 1.23 × 10−2 |
| LOC105196698 | probable salivary secreted peptide | 463.02 | 1.66 | −8.12 | 1.60 × 10−3 | 4.76 × 10−2 |
| LOC105199102 | venom metalloproteinase 3-like | 240.80 | 1.27 | −7.57 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105196175 | zinc carboxypeptidase-like | 2455.41 | 18.21 | −7.08 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105193306 | chymotrypsin-1-like | 186.87 | 1.76 | −6.73 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105196420 | peritrophin-1-like | 1162.08 | 15.00 | −6.28 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200093 | chymotrypsin-2-like, partial | 639.60 | 10.38 | −5.95 | 1.50 × 10−4 | 6.96 × 10−3 |
| LOC105197108 | aminopeptidase N, partial | 15.65 | 0.26 | −5.91 | 1.50 × 10−4 | 6.96 × 10−3 |
| LOC105205908 | chymotrypsin-2-like, partial | 216.51 | 4.93 | −5.46 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105207761 | anionic trypsin-2-like | 100.83 | 3.36 | −4.91 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200277 | inducible metalloproteinase inhibitor protein-like | 27.20 | 1.03 | −4.72 | 6.50 × 10−4 | 2.28 × 10−2 |
| LOC105200273 | chymotrypsin inhibitor-like | 1284.77 | 56.33 | −4.51 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200221 | retinoid-inducible serine carboxypeptidase-like | 169.37 | 7.85 | −4.43 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200003 | glutamyl aminopeptidase isoform X3 | 123.56 | 6.06 | −4.35 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105193273 | chymotrypsin-2-like | 156.98 | 8.14 | −4.27 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105195679 | serine proteinase stubble | 35.89 | 2.16 | −4.05 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105203057 | endoplasmic reticulum metallopeptidase 1-like isoform X4 | 21.71 | 1.57 | −3.79 | 1.50 × 10−4 | 6.96 × 10−3 |
| LOC105205350 | xaa-Pro dipeptidase | 103.15 | 7.71 | −3.74 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200222 | retinoid-inducible serine carboxypeptidase-like | 97.19 | 7.46 | −3.70 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105203057 | endoplasmic reticulum metallopeptidase 1-like isoform X2 | 81.14 | 7.32 | −3.47 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105206478 | aminopeptidase N-like, partial | 107.22 | 10.36 | −3.37 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105203676 | dipeptidyl peptidase 3 isoform X2 | 189.57 | 19.73 | −3.26 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200810 | digestive cysteine proteinase 1 | 1,995.67 | 217.85 | −3.20 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105206533 | aminopeptidase N-like, partial | 122.02 | 13.78 | −3.15 | 1.00 × 10−4 | 4.96 × 10−3 |
| LOC105203240 | trehalase-like | 25.37 | 3.41 | −2.90 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105196148 | cytosolic non-specific dipeptidase | 217.85 | 34.02 | −2.68 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105195467 | xaa-Pro aminopeptidase 1 | 95.29 | 15.49 | −2.62 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105196184 | signal peptidase complex subunit 1 | 98.78 | 20.91 | −2.24 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105204623 | mitochondrial-processing peptidase subunit beta | 163.92 | 35.95 | −2.19 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105197517 | aminopeptidase N | 11.48 | 2.80 | −2.04 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105198213 | signal peptidase complex subunit 3 | 300.89 | 74.52 | −2.01 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105199614 | puromycin-sensitive aminopeptidase isoform X1 | 61.68 | 16.73 | −1.88 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200356 | signal peptidase complex catalytic subunit SEC11A | 73.13 | 20.48 | −1.84 | 5.00 × 10−4 | 1.85 × 10−2 |
| LOC105198128 | probable signal peptidase complex subunit 2 | 100.86 | 33.86 | −1.57 | 1.00 × 10−4 | 4.96 × 10−3 |
| LOC105193333 | prolyl endopeptidase-like, partial | 87.30 | 32.62 | −1.42 | 6.00 × 10−4 | 2.14 × 10−2 |
| LOC105201160 | mitochondrial-processing peptidase subunit alpha | 51.37 | 20.68 | −1.31 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105201089 | putative phospholipase B-like lamina ancestor | 214.83 | 101.38 | −1.08 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105200792 | disintegrin and metalloproteinase with thrombospondin motifs 7-like | 3.97 | 11.35 | 1.52 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105197096 | peritrophin-1-like, partial | 13.82 | 45.98 | 1.73 | 1.00 × 10−4 | 4.96 × 10−3 |
| LOC105193940 | disintegrin and metalloproteinase domain-containing protein 11, partial | 2.61 | 10.41 | 2.00 | 6.00 × 10−4 | 2.14 × 10−2 |
| LOC105193191 | sn1-specific diacylglycerol lipase beta-like | 2.34 | 10.15 | 2.12 | 4.50 × 10−4 | 1.70 × 10−2 |
| LOC105205498 | dipeptidase 1-like | 1.02 | 5.77 | 2.50 | 1.05 × 10−3 | 3.38 × 10−2 |
| LOC105202515 | salivary plasminogen activator gamma | 4.49 | 30.83 | 2.78 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105202515 | salivary plasminogen activator gamma | 1.69 | 22.46 | 3.73 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105197409 | membrane metallo-endopeptidase-like 1 isoform X4 | 5.05 | 71.25 | 3.82 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105198885 | serine proteinase stubble | 0.71 | 22.38 | 4.98 | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105194705 | carboxypeptidase B-like | 4.20 | 145.31 | 5.11 | 8.50 × 10−4 | 2.84 × 10−2 |
| LOC105198972 | phospholipase B1, membrane-associated-like, partial | - | 1.83 | inf | 5.00 × 10−5 | 2.75 × 10−3 |
| LOC105205146 | phospholipase B1, membrane-associated-like, partial | - | 2.47 | inf | 5.00 × 10−5 | 2.75 × 10−3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allen, M.L.; Rhoades, J.H.; Sparks, M.E.; Grodowitz, M.J. Differential Gene Expression in Red Imported Fire Ant (Solenopsis invicta) (Hymenoptera: Formicidae) Larval and Pupal Stages. Insects 2018, 9, 185. https://doi.org/10.3390/insects9040185
Allen ML, Rhoades JH, Sparks ME, Grodowitz MJ. Differential Gene Expression in Red Imported Fire Ant (Solenopsis invicta) (Hymenoptera: Formicidae) Larval and Pupal Stages. Insects. 2018; 9(4):185. https://doi.org/10.3390/insects9040185
Chicago/Turabian StyleAllen, Margaret L., Joshua H. Rhoades, Michael E. Sparks, and Michael J. Grodowitz. 2018. "Differential Gene Expression in Red Imported Fire Ant (Solenopsis invicta) (Hymenoptera: Formicidae) Larval and Pupal Stages" Insects 9, no. 4: 185. https://doi.org/10.3390/insects9040185
APA StyleAllen, M. L., Rhoades, J. H., Sparks, M. E., & Grodowitz, M. J. (2018). Differential Gene Expression in Red Imported Fire Ant (Solenopsis invicta) (Hymenoptera: Formicidae) Larval and Pupal Stages. Insects, 9(4), 185. https://doi.org/10.3390/insects9040185