Salmonella enterica Subsp. houtenae Associated with an Abscess in Young Roe Deer (Capreolus capreolus)
Abstract
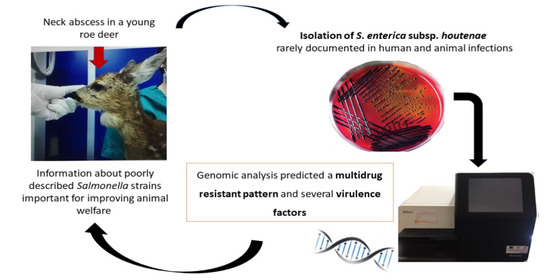
:1. Introduction
2. Case Report
3. Results
3.1. Microbiological Investigation
3.2. Genomic Analysis
4. Discussion
5. Material and Methods
5.1. Microbiological Investigation
5.2. Genomic Analysis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zottola, T.; Montagnaro, S.; Magnapera, C.; Sasso, S.; De Martino, L.; Bragagnolo, A.; D’Amici, L.; Condoleo, R.; Pisanelli, G.; Iovane, G.; et al. Prevalence and antimicrobial susceptibility of Salmonella in European wild boar (Sus scrofa); Latium Region—Italy. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 161–168. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar]
- Sandala, J.; Gunn, J.S. In Vitro Evaluation of Anti-biofilm Agents Against Salmonella enterica. Methods Mol. Biol. 2020, 2182, 127–139. [Google Scholar] [CrossRef]
- Lamas, A.; Miranda, J.M.; Regal, P.; Vazquez, B.; Franco, C.M.; Cepeda, A. A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiol. Res. 2018, 206, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Handeland, K.; Refsum, T.; Johansen, B.S.; Holstad, G.; Knutsen, G.; Solberg, I.; Schulze, J.; Kapperud, G. Prevalence of Salmonella typhimurium infection in Norwegian hedgehog populations associated with two human disease outbreaks. Epidemiol. Infect. 2002, 128, 523–527. [Google Scholar] [CrossRef]
- Millan, J.; Aduriz, G.; Moreno, B.; Juste, R.; Barral, M. Salmonella isolates from wild birds and mammals in the Basque Country (Spain). Rev. Sci. Tech. OIE 2004, 23, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Corrente, M.; Madio, A.; Friedrich, K.; Greco, G.; Desario, C.; Tagliabue, S.; D’Incau, M.; Campolo, M.; Buonavoglia, C. Isolation of Salmonella strains from reptile faeces and comparison of different culture media. J. Appl. Microbiol. 2004, 96, 709–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrente, M.; Sangiorgio, G.; Grandolfo, E.; Bodnar, L.; Catella, C.; Trotta, A.; Martella, V.; Buonavoglia, D. Risk for zoonotic Salmonella transmission from pet reptiles: A survey on knowledge, attitudes and practices of reptile-owners related to reptile husbandry. Prev. Vet. Med. 2017, 146, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Renter, D.G.; Gnad, D.P.; Sargeant, J.M.; Hygnstrom, S.E. Prevalence and Serovars of Salmonella in the Feces of Free-Ranging White-Tailed Deer (Odocoileus virginianus) in Nebraska. J. Wildl. Dis. 2006, 42, 699–703. [Google Scholar] [CrossRef] [Green Version]
- Foreyt, W.J.; Besser, T.E.; Lonning, S.M. Mortality in Captive Elk from Salmonellosis. J. Wildl. Dis. 2001, 37, 399–402. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Kobayashi, C.; Ichikawa, K.; Kuwamoto, R.; Matsuura, S.; Koyama, T. An Occurrence of Salmonella Typhimurium Infection in Sika Deer (Cervus nippon). J. Vet. Med. Sci. 2000, 62, 313–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grandolfo, E.; Parisi, A.; Ricci, A.; Lorusso, E.; De Siena, R.; Trotta, A.; Buonavoglia, D.; Martella, V.; Corrente, M. High mortality in foals associated with Salmonella enterica subsp. enterica Abortusequi infection in Italy. J. Vet. Diagn. Investig. 2018, 30, 483–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küker, U.S.; Huber, N.; Evans, A.; Kjellander, P.; Bergvall, U.A.; Jones, K.L.; Arnemo, J.M. Hematology, Serum Chemistry, and Serum Protein Electrophoresis Ranges for Free-ranging Roe Deer (Capreolus capreolus) in Sweden. J. Wildl. Dis. 2015, 51, 269–273. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Chen, C.-C.; Chiou, C.-S. Construction of a Pan-Genome Allele Database of Salmonella enterica Serovar Enteritidis for Molecular Subtyping and Disease Cluster Identification. Front. Microbiol. 2016, 7, 2010. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Pellegrini-Masini, A.; Dolente, B.A.; Habecker, P.L.; Jesty, S.A. Myonecrosis and cutaneous infarction associated with Salmonella serovar Infantum infection in a horse. J. Am. Vet. Med. Assoc. 2004, 225, 722–725. [Google Scholar] [CrossRef]
- Tavechio, A.; Ghilardi, A.; Peresi, J.; Fuzihara, T.; Yonamine, E.; Jakabi, M.; Fernandes, S. Serotypes Isolated from Nonhuman Sources in São Paulo, Brazil, from 1996 through 2000. J. Food Prot. 2002, 65, 1041–1044. [Google Scholar] [CrossRef]
- Abbott, S.L.; Ni, F.C.; Janda, J.M. Increase in Extraintestinal Infections Caused by Salmonella enterica Subspecies II–IV. Emerg. Infect. Dis. 2012, 18, 637–639. [Google Scholar] [CrossRef]
- Franco, A.; Hendriksen, R.S.; Lorenzetti, S.; Onorati, R.; Gentile, G.; Dell’Omo, G.; Aarestrup, F.M.; Battisti, A. Characterization of Salmonella Occurring at High Prevalence in a Population of the Land Iguana Conolophus subcristatus in Galápagos Islands, Ecuador. PLoS ONE 2011, 6, e23147. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.R.; Singh, V.; Ebibeni, N.; Singh, R.K. Antimicrobial and Herbal Drug Resistance in Enteric Bacteria Isolated from Faecal Droppings of Common House Lizard/Gecko (Hemidactylus frenatus). Int. J. Microbiol. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu-Kee, E.; Priyia, P.; Nurul-Syakima Ab, M.; Hooi-Leng, S.; Kok-Gan, C.; Learn-Han, L. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar]
- Hyeon, J.Y.; Li, S.; Mann, D.A.; Zhang, S.; Kim, K.J.; Lee, D.H.; Deng, X.; Song, C.S. Whole-Genome Sequencing Analysis of Salmonella enterica Serotype Enteritidis Isolated from Poultry Sources in South Korea, 2010–2017. Pathogens 2021, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.L.; Brumell, J.H.; Pfeifer, C.G.; Finlay, B. Salmonella pathogenicity islands: Big virulence in small packages. Microbes Infect. 2000, 2, 145–156. [Google Scholar] [CrossRef]
- Diniz, J.A.; Liu, Y.; Coulthurst, S.J. Molecular weaponry: Diverse effectors delivered by the Type VI secretion system. Cell. Microbiol. 2015, 17, 1742–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Asten, A.J.A.M.; van Dijk, J.E. Distribution of “classic” virulence factors among Salmonella spp. FEMS Microbiol. Rev. 2005, 44, 251–259. [Google Scholar]
- Hensel, M. Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 2004, 294, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Blanc-Potard, A.; Groisman, E.A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997, 16, 5376–5385. [Google Scholar] [CrossRef]
- Wallis, T.S.; Wood, M.; Watson, P.; Paulin, S.; Jones, M.; Galyov, E. SIPS, SOPS, and SPIs but not STN Influence Salmonella Enteropathogenesis. Adv. Exp. Med. Biol. 1999, 473, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.W.; Rosqvist, R.; Mullan, P.B.; Edwards, M.H.; Galyov, E.E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 1996, 22, 327–338. [Google Scholar] [CrossRef]
- Rushing, M.D.; Slauch, J.M. Either periplasmic tethering or protease resistance is sufficient to allow a SodC to protect Salmonella enterica serovar Typhimurium from phagocytic superoxide. Mol. Microbiol. 2011, 82, 952–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moncrief, M.B.C.; Maguire, M.E. Magnesium and the Role of mgtC in Growth of Salmonella typhimurium. Infect. Immun. 1998, 66, 3802–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoo, C.-H.; Sim, J.-H.; Salleh, N.A.; Cheah, Y.-K. Pathogenicity and phenotypic analysis of sopB, sopD and pipD virulence factors in Salmonella enterica serovar typhimurium and Salmonella enterica serovar Agona. Antonie Van Leeuwenhoek 2014, 107, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 1996, 20, 263–271. [Google Scholar] [CrossRef]
- Ganesan, V.; Harish, B.N.; Menezes, G.A.; Parija, S.C. Detection of Salmonella in Blood by PCR using iroB gene. J. Clin. Diagn. Res. 2014, 8, DC01–DC03. [Google Scholar] [CrossRef]
- Hantke, K.; Nicholson, G.; Rabsch, W.; Winkelmann, G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 2003, 100, 3677–3682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiya, K.-I.; Nikai, T. Salmonella virulence factor SpiC is involved in expression of flagellin protein and mediates activation of the signal transduction pathways in macrophages. Microbiology 2008, 154, 3491–3502. [Google Scholar] [CrossRef] [Green Version]
- Kolenda, R.; Ugorski, M.; Grzymajlo, K. Everything You Always Wanted to Know About Salmonella Type 1 Fimbriae, but were Afraid to Ask. Front. Microbiol. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Olsen, J.E.; Hoegh-Andersen, K.H.; Casadesús, J.; Rosenkranzt, J.; Chadfield, M.S.; Thomsen, L.E. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 2013, 13, 67. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.A.; Nawaz, M.S.; Khan, S.A.; Cerniglia, C.E. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 2000, 182, 355–360. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Bianco, A.; Capozzi, L.; Monno, M.R.; Del Sambro, L.; Manzulli, V.; Pesole, G.; Loconsole, D.; Parisi, A. Characterization of Bacillus cereus Group Isolates from Human Bacteremia by Whole-Genome Sequencing. Front. Microbiol. 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiden, M.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmo nella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 22, e0147101. [Google Scholar]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the NCBI AMRFinder tool and resistance gene database using antimicrobial resistance genotype-phenotype correlations in a collection of NARMS isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Kaas, R.S.; Seyfarth, A.M.; Agersø, Y.; Lund, O.; Larsen, M.V.; Aarestrup, F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 771–777. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, C.; Laird, M.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. Simon Fraser University Research Computing Group IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
| Class | Number of Genes | Function |
|---|---|---|
| J * | 245 | Translation, ribosomal structure, and biogenesis |
| A | 25 | RNA processing and modification |
| K | 231 | Transcription |
| L | 238 | Replication, recombination, and repair |
| B | 19 | Chromatin structure and dynamics |
| D | 72 | Cell cycle control, cell division, and chromosome partitioning |
| Y | 2 | Nuclear structure |
| V | 46 | Defense mechanisms |
| T | 152 | Signal transduction mechanisms |
| M | 188 | Cell wall/membrane/envelope biogenesis |
| N | 96 | Cell motility |
| Z | 12 | Cytoskeleton |
| W | 1 | Extracellular structures |
| U | 158 | Intracellular trafficking, secretion, and vesicular transport |
| O | 203 | Posttranslational modification, protein turnover, and chaperones |
| C | 258 | Energy production and conversion |
| G | 230 | Carbohydrate transport and metabolism |
| E | 270 | Amino acid transport and metabolism |
| F | 95 | Nucleotide transport and metabolism |
| H | 179 | Coenzyme transport and metabolism |
| I | 94 | Lipid transport and metabolism |
| P | 212 | Inorganic ion transport and metabolism |
| Q | 88 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 702 | General function prediction only |
| S | 1347 | Unknown |
| Aminoglycoside | Multidrug | Fluoroquinolone | Nitroimidazole | Peptide | Fosfomycin | Aminocoumarin | ß-Lactam | Tetracycline | |
|---|---|---|---|---|---|---|---|---|---|
| AAC(6′)-Iy | ampH | LptD | emrA | msbA | arnA | mdtG | mdtA | (Bla)AMPH E. coli | tet(34) |
| acrD | acrA | marA | emrB | ArnT | mdtB | (Bla)PBP E. coli | |||
| kdpE | acrB | mdfA | emrR | bacA | mdtC | ||||
| acrE | mdtM | mdtH | eptA | ||||||
| acrF | mdtN | mdtK | eptB | ||||||
| acrS | mdtO | yojI | |||||||
| baeS | mdtP | pmrF | |||||||
| baeR | OmpA | rosA | |||||||
| cpxA | OmpK37 | rosB | |||||||
| CRP | ramA | ugd | |||||||
| golS | rsmA | ||||||||
| KpnE | sdiA | ||||||||
| KpnF | tolC | ||||||||
| H-NS | |||||||||
| Virulence Factors | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adherence | Magnesium Uptake | Secretion System | Stress Protein | Toxin | Pathogenicity Islands | |||||||
| Agf | Type 1 fimbriae | MgtBC | TTSS (SPI-1 encode) | TTSS (SPI-2 encode) | Type VI effector | sodCI | spvB | SPI-1 | SPI-2 | SPI-5 | ||
| csgA | fimC | mgtB | invA | pipB2 | AHA_1833 | invA | sicA | spiC/ssaB | ssaR | pipB2 | ||
| csgB | fimD | mgtC | invE | sifA | fha | invB | sicP | ssaC | ssaS | sopB | ||
| csgC | fimF | invF | sifB | galF | invC | sipA/sspA | ssaD | ssaT | ||||
| csgD | fimH | invG | sopD2 | hcp2/tssD2 | invE | sipB/sspB | ssaE | ssaU | ||||
| csgE | fimI | invH | spiC/ssaB | tssA | invF | sipC/sspC | ssaG | ssaV | ||||
| csgG | invI | sseA | tssB | invG | sipD | ssaH | sscA | |||||
| invJ | ssaD | tssJ | invH | spaO | ssaI | sscB | ||||||
| orgA/sctK | sscA | tssL | invI | spaP | ssaJ | sseA | ||||||
| prgH | ssaC | tssM | invJ | spaQ | ssaK | sseB | ||||||
| prgI | sscB | rhs/PAAR | orgA | spaR | ssaL | sseC | ||||||
| prgJ | sseB | orgB | spaS | ssaM | sseD | |||||||
| prgK | sseC | orgC | ssaN | sseE | ||||||||
| sicA | sseD | ssaO | sseF | |||||||||
| sipB/sspB | sseE | ssaP | sseG | |||||||||
| spaO/sctQ | sseF | ssaQ | ||||||||||
| spaP | sseG | |||||||||||
| spaQ | sspH1 | |||||||||||
| spaR | sspH2 | |||||||||||
| spaS | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trotta, A.; Del Sambro, L.; Galgano, M.; Ciccarelli, S.; Ottone, E.; Simone, D.; Parisi, A.; Buonavoglia, D.; Corrente, M. Salmonella enterica Subsp. houtenae Associated with an Abscess in Young Roe Deer (Capreolus capreolus). Pathogens 2021, 10, 654. https://doi.org/10.3390/pathogens10060654
Trotta A, Del Sambro L, Galgano M, Ciccarelli S, Ottone E, Simone D, Parisi A, Buonavoglia D, Corrente M. Salmonella enterica Subsp. houtenae Associated with an Abscess in Young Roe Deer (Capreolus capreolus). Pathogens. 2021; 10(6):654. https://doi.org/10.3390/pathogens10060654
Chicago/Turabian StyleTrotta, Adriana, Laura Del Sambro, Michela Galgano, Stefano Ciccarelli, Erika Ottone, Domenico Simone, Antonio Parisi, Domenico Buonavoglia, and Marialaura Corrente. 2021. "Salmonella enterica Subsp. houtenae Associated with an Abscess in Young Roe Deer (Capreolus capreolus)" Pathogens 10, no. 6: 654. https://doi.org/10.3390/pathogens10060654
APA StyleTrotta, A., Del Sambro, L., Galgano, M., Ciccarelli, S., Ottone, E., Simone, D., Parisi, A., Buonavoglia, D., & Corrente, M. (2021). Salmonella enterica Subsp. houtenae Associated with an Abscess in Young Roe Deer (Capreolus capreolus). Pathogens, 10(6), 654. https://doi.org/10.3390/pathogens10060654