Competing Bioaerosols May Influence the Seasonality of Influenza-Like Illnesses, including COVID-19. The Chicago Experience
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Counting of Pollens and Mold Spores
2.2. ILI and COVID-19 Data
2.3. Data Analysis
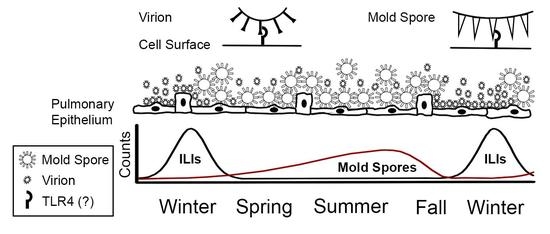
3. Results
3.1. Kinetics of Presentations to Emergency Departments
3.2. Kinetics of Pollen and Mold Spore Counts
3.3. Inhibition of ILI and COVID-19 Presentations by Mold Spores
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lofgren, E.; Fefferman, N.H.; Naumov, Y.N.; Gorski, J.; Naumova, E.N. Influenza Seasonality: Underlying Causes and Modeling Theories. J. Virol. 2007, 81, 5429–5436. [Google Scholar] [CrossRef] [Green Version]
- Monto, A.S.; DeJonge, P.M.; Callear, A.P.; Bazzi, L.A.; Capriola, S.B.; Malosh, R.E.; Martin, E.T.; Petrie, J.G. Coronavirus Occurrence and Transmission over 8 Years in the HIVE Cohort of Households in Michigan. J. Infect. Dis. 2020, 222, 9–16. [Google Scholar] [CrossRef]
- Hoogeveen, M.J.; van Gorp, E.C.M.; Hoogeveen, E.K. Pollen Explains Flu-like and COVID-19 Seasonality. medRxiv 2020. [Google Scholar] [CrossRef]
- Hoogeveen, M.J. Pollen Likely Seasonal Factor in Inhibiting Flu-like Epidemics. A Dutch Study into the Inverse Relation between Pollen Counts, Hay Fever and Flu-like Incidence 2016–2019. Sci. Total Environ. 2020, 727, 138543. [Google Scholar] [CrossRef]
- Kanter, U.; Heller, W.; Durner, J.; Winkler, J.B.; Engel, M.; Behrendt, H.; Holzinger, A.; Braun, P.; Hauser, M.; Ferreira, F.; et al. Molecular and Immunological Characterization of Ragweed (Ambrosia artemisiifolia L) Pollen after Exposure of the Plants to Elevated Ozone over a Whole Growing Season. PLoS ONE 2013, 8, e61518. [Google Scholar] [CrossRef]
- Martinez, A.T.; Calvo, M.A.; Ramirez, C. Scanning Electron Microscopy of Penicillium Conidia. Antonie Leeuwenhoek 1982, 48, 245–255. [Google Scholar] [CrossRef]
- Sannier, J.; Baker, W.J.; Anstett, M.-C.; Nadot, S. A Comparative Analysis of Pollinator Type and Pollen Ornamentation in the Araceae and the Arecaceae, Two Unrelated Families of the Monocots. BMC Res. Notes 2009, 2, 145. [Google Scholar] [CrossRef] [Green Version]
- Silva, D.M.; Batista, L.R.; Rezende, E.F.; Fungaro, M.H.P.; Sartori, D.; Alves, E. Identification of Fungi of the Genus Aspergillus Section nigri Using Polyphasic Taxonomy. Braz. J. Microbiol. 2011, 42, 761–773. [Google Scholar] [CrossRef] [Green Version]
- Dijksterhuis, J.; van Egmond, W.; Yarwood, A. From Colony to Rodlet. A Six Meter Long Portrait of the Xerophilic Fungus Aspergillus restrictus Decorates the Hall of the Westerdijk Institute. Fungal Biol. 2020, 124, 509–515. [Google Scholar] [CrossRef]
- Paris, S.; Debeaupuis, J.-P.; Crameri, R.; Carey, M.; Charlès, F.; Prévost, M.C.; Schmitt, C.; Philippe, B.; Latgé, J.P. Conidial Hydrophobins of Aspergillus fumigatus. Appl. Environ. Micro. 2003, 69, 1581–1588. [Google Scholar] [CrossRef] [Green Version]
- Kurtzman, C.P.; Smiley, M.J.; Baker, F.L. Scanning Electron Microscopy of Ascospores of Schwanniomyces. J. Bacteriol. 1972, 112, 1380–1382. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probably Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Sohn, K.M.; Lee, S.-G.; Kim, H.J.; Cheon, S.; Jeong, H.; Lee, J.; Kim, I.S.; Silwal, P.; Kim, Y.J.; Paik, S.; et al. COVID-19 Patients Upregulate Toll-like Receptor 4-Mediated Inflammatory Signaling that Mimics Bacterial Sepsis. J. Korean Med. Sci. 2020, 35, e343. [Google Scholar] [CrossRef]
- Aboudounya, M.M.; Heads, R.J. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Med. Inflamm. 2021, 2021, 8874339. [Google Scholar] [CrossRef]
- Nhu, Q.M.; Shirey, K.; Teijaro, J.R.; Farber, D.; Netzel-Arnett, S.; Antalis, T.M.; Fasano, A.; Vogel, S.N. Novel Signaling Interactions between Proteinase-Activated Receptor 2 and Toll-like Receptors In vitro and In vivo. Mucosal Immunol. 2010, 3, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Shirey, K.A.; Lai, W.; Brown, L.J.; Blanco, J.C.G.; Beadenkopf, R.; Wang, Y.; Vogel, S.N.; A Snyder, G. Select Targeting of Intracellular Toll-Interleukin-1 Receptor Resistance Domains for Protection Against Influenza-Induced Disease. Innate Immun. 2020, 26, 26–34. [Google Scholar] [CrossRef]
- Shirey, K.a.; Lai, W.; Patel, M.C.; Pletneva, L.M.; Pang, C.; Kurt-Jones, E.; Lipsky, M.; Roger, T.; Calandra, T.; Tracey, K.J.; et al. Novel Strategies for Targeting Innate Immune Responses to Influenza. Mucosal Immunol. 2016, 9, 1173–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, T.; Rumore, A.; Howard, B.; He, X.; Luo, M.; Wuenschmann, S.; Chapman, M.; Kale, S.; Li, L.; Kita, H.; et al. Innate Immunity Induced by the Major Allergen Alt a 1 from the Fungus Alternaria is Dependent upon Toll-like Receptors 2/4 in Human Lung Epithelial Cells. Front. Immunol. 2018, 9, 1507. [Google Scholar] [CrossRef] [Green Version]
- Roeder, A.; Kirschning, C.J.; Rupec, R.A.; Schaller, M.; Weindl, G.; Korting, H.C. Toll-like Receptors as Key Mediators in Innate Antifungal Immunity. Med. Mycol. 2004, 42, 485–498. [Google Scholar] [CrossRef]
- Francone, M.; Iafrate, F.; Masci, G.M.; Coco, S.; Cilia, F.; Manganaro, L.; Panebianco, V.; Andreoli, C.; Colaiacomo, M.C.; Zingaropoli, M.A.; et al. Score in COVID-19 Patients: Correlation with Disease Severity and Short-term Prognosis. Eur. Radiol. 2020, 30, 1–10. [Google Scholar] [CrossRef]
- Xiao, J.; Li, X.; Xie, Y.; Huang, Z.; Ding, Y.; Zhao, S.; Yang, P.; Du, D.; Liu, B.; Wang, X. Maximum Chest CT Score is Associated with Progression to Severe Illness in Patients with COVID-19: A Retrospective Study from Wuhan, China. BMC Infect. Dis. 2020, 20, 953. [Google Scholar] [CrossRef]
- Armstrong, L.; Medford, A.R.L.; Uppington, K.M.; Robertson, J.; Witherden, I.R.; Teresa, T.D.; Miller, A.B. Expression of Functional Toll-like Receptor-2 and -4 on Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2004, 31, 241–245. [Google Scholar] [CrossRef]
- Hearps, A.C.; Martin, G.E.; Angelovich, T.; Cheng, W.-J.; Maisa, A.; Landay, A.L.; Jaworowski, A.; Crowe, S.M. Aging is Associated with Chronic Immune Activation and Dysregulation of Monocyte Phenotype and Function. Aging Cell 2012, 11, 867–875. [Google Scholar] [CrossRef]
- Hussain, S.; Johnson, C.G.; Sciurba, J.; Meng, X.; Stober, V.P.; Liu, C.; Cyphert-Daly, J.M.; Bulek, K.; Qian, W.; Solis, A.; et al. TLR5 Participates in the TLR4 Receptor Complex and Promotes MyD88-Dependent Signaling in Environmental Lung Injury. eLife 2020, 9, e50458. [Google Scholar] [CrossRef]
- Qian, F.; Wang, X.; Zhang, L.; Chen, S.; Piecychna, M.; Allore, H.; Bockenstedt, L.; Malawista, S.; Bucala, R.; Shaw, A.C.; et al. Age-Associated Elevation in TLR5 Leads to Increased Inflammatory Responses in the Elderly. Aging Cell 2012, 11, 104–119. [Google Scholar] [CrossRef]
- Zuliani-Alvarez, L.; Marzeda, A.; Deligne, C.; Schwenzer, A.; McCann, F.E.; Marsden, B.D.; Piccinini, A.M.; Midwood, K.S. Mapping Tenascin-C Interaction with Toll-like Receptor 4 Reveals a New Subset of Endogenous Inflammatory Triggers. Nat. Comm. 2017, 8, 1595. [Google Scholar] [CrossRef]
- Hardy, M.; Michaux, I.; Lessire, S.; Douxfils, J.; Dogné, J.-M.; Bareille, M.; Horlait, G.; Bulpa, P.; Chapelle, C.; Laporte, S.; et al. Prothrombotic Disturbances of Hemostasis of Patients with Severe COVID-19: A Prospective Longitudinal Observational Study. Thromb. Res. 2021, 197, 20–23. [Google Scholar] [CrossRef]
- Hu, W.; Xie, J.; Chau, H.W.; Si, B.C. Evaluation of Parameter Uncertainties in Nonlinear Regression Using Microsoft Excel Spreadsheet. Environ. Syst. Res. 2015, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- McKinnon, G.H.; Backhouse, C.J.; Kalantar, A.H. Optimizing the use of the Kézdy-Mangelsdorf-Swinbourne Method for Analysis of Data Following A exp(-kt) + Z. Int. J. Chem. Kinet. 1984, 16, 1427–1454. [Google Scholar] [CrossRef]
- Retzinger, D.G.; Retzinger, A.C.; Retzinger, G.R. Estimate of Benefit Attributable to Wearing Masks in Chicago during the Early Days of the Pandemic. Med. Hypotheses 2021, in press. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [Green Version]
- Noman, A.; Aqeel, M.; Qasim, M.; Haider, I.; Lou, Y. Plant-Insect-Microbe Interaction: A Love Triangle between Enemies in Ecosystem. Sci. Total Environ. 2020, 699, 134181. [Google Scholar] [CrossRef]
- Biedermann, P.H.W.; Vega, F.E. Ecology and Evolution of Insect—Fungus Mutualisms. Annu. Rev. Entomol. 2020, 65, 431–455. [Google Scholar] [CrossRef] [Green Version]
- Kevan, P.G.; Baker, H.G. Insects as Flower Visitors and Pollinators. Annu. Rev. Entomol. 1983, 28, 407–453. [Google Scholar] [CrossRef]
- Rader, R.; Cunningham, S.A.; Howlett, B.G.; Inouye, D.W. Non-Bee Insects as Visitors and Pollinators of Crops: Biology, Ecology, and Management. Annu. Rev. Entomol. 2020, 65, 391–407. [Google Scholar] [CrossRef] [Green Version]
- da Silva, L.P.; Coutinho, A.P.; Heleno, R.H.; Tenreiro, P.Q.; Ramos, J.A. Dispersal of Fungi Spores by Non-Specialized Flower-Visiting Birds. J. Avian Biol. 2016, 47, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Muchhala, N.; Thomson, J.D. Fur versus Feathers: Pollen Delivery by Bats and Hummingbirds and Consequences for Pollen Production. Am. Nat. 2010, 75, 717–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diniz, U.M.; Lima, S.A.; Machado, I.C.S. Short-Distance Pollen Dispersal by Bats in an Urban Setting: Monitoring the Movement of Vertebrate Pollinator through Fluorescent Dyes. Urban Ecosyst. 2019, 22, 281–291. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. Ectomycota Associated with Arthropods from Bat Hibernacula in Eastern Canada, with Particular Reference to Pseudogymnoascus destructans. Insects 2016, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subudhi, S.; Rapin, N.; Misra, V. Immune System Modulation and Viral Persistence in Bats: Understanding Viral Spillover. Viruses 2019, 11, 192. [Google Scholar] [CrossRef] [Green Version]
- Linnakoski, R.; Reshamwala, D.; Veteli, P.; Cortina-Escribano, M.; Vanhanen, H.; Marjomäki, V. Antiviral Agents from Fungi: Diversity, Mechanisms and Potential Applications. Front. Microbiol. 2018, 9, 2325. [Google Scholar] [CrossRef] [Green Version]
- Pobiega, K.; Gniewosz, M.; Kraśniewska, K. Antimicrobial and Antiviral Properties of Different Types of Propolis. Zesz. Probl. Postępów Nauk. Rol. 2017, 589, 69–79. [Google Scholar] [CrossRef]
- Van Kerkhove, M.D.; Vandemaele, K.A.H.; Shinde, V.; Jaramillo-Gutierrez, G.; Koukounari, A.; Donnelly, C.A.; Carlino, L.O.; Owen, R.; Paterson, B.; Pelletier, L.; et al. Risk Factors for Severe Outcomes Following 2009 Influenza A (H1N1) Infection: A Global Pooled Analysis. PLoS Med. 2011, 8, e1001053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Cao, J.; Yu, Y.; Ding, J.; Eshak, E.S.; Liu, K.; Mubarik, S.; Shi, F.; Wen, H.; Zeng, Z.; et al. Epidemiological Characteristics of Patients with Severe COVID-19 Infection in Wuhan, China: Evidence from a Retrospective Observational Study. Int. J. Epidemiol. 2020, 49, 1940–1950. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.A.; Waite, B.; Thompson, M.G.; McArthur, C.; Wong, C.; Baker, M.G.; Wood, T.; Haubrock, J.; Roberts, S.; Gross, D.K.; et al. Risk of Severe Influenza among Adults with Chronic Medical Conditions. J. Infect. Dis. 2020, 221, 183–190. [Google Scholar] [CrossRef]
- Chhiba, K.D.; Patel, G.B.; Vu, T.H.T.; Chen, M.M.; Guo, A.; Kudlaty, E.; Mai, Q.; Yeh, C.; Muhammad, L.N.; Harris, K.E.; et al. Prevalence and Characterization of Asthma in Hospitalized and Nonhospitalized Patients with COVID-19. J. Allergy Clin. Immunol. 2020, 146, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, F.; Keshavarzi, A.; Akbari, A.; Emami, A.; Pirbonyeh, N. Prevalence of Underlying Diseases in Died Cases of COVID-19: A Systematic Review and Meta-analysis. PLoS ONE 2020, 15, e0241265. [Google Scholar] [CrossRef]
- Choudhury, A.; Mukherjee, S. In silico Studies on the Comparative Characterization of the Interactions of SARS-CoV-2 Spike Glycoprotein with ACE-2 Receptor Homologs and Human TLRs. J. Med. Virol. 2020, 92, 1–9. [Google Scholar] [CrossRef]
- Koldehoff, M.; Beelen, D.W.; Elmaagacli, A.H. Increased Susceptibility for Aspergillosis and Post-Transplant Immune Deficiency in Patient with Gene Variants of TLR4 after Stem Cell Transplantation. Transpl. Infect. Dis. 2013, 15, 533–539. [Google Scholar] [CrossRef]
- Carvalho, A.; Pasqualotto, A.C.; Pitzurra, L.; Romani, L.; Denning, D.W.; Rodrigues, F. Polymorphisms in Toll-like Receptor Genes and Susceptibility to Pulmonary Aspergillosis. J. Infect. Dis. 2008, 197, 618–621. [Google Scholar] [CrossRef] [Green Version]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The Protein Expression Profile of ACE2 in Human Tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef]
- Lee, I.T.; Nakayama, T.; Wu, C.-T.; Goltsev, Y.; Jiang, S.; Gall, P.A.; Liao, C.-K.; Shih, L.-C.; Schürch, C.M.; McIlwain, D.R.; et al. ACE2 Localizes to the Respiratory Cilia and is Not Increased by ACE Inhibitors or ARBs. Nat. Commun. 2020, 11, 5453. [Google Scholar] [CrossRef]
- Lee, N.; Wong, C.K.; Hui, D.; Lee, S.K.W.; Wong, R.Y.K.; Ngai, K.L.K.; Chan, M.C.-W.; Chu, Y.J.; Ho, A.W.Y.; Lui, C.Y.G.; et al. Role of Human Toll-like Receptors in Naturally Occurring Influenza A Infections. Influenza Other Respir. Viruses 2013, 7, 666–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchant, D.; Singhera, G.K.; Utokaparch, S.; Hackett, T.L.; Boyd, J.H.; Luo, Z.; Si, X.; Dorscheid, D.R.; McManus, B.M.; Hegele, R.G. Toll-like Receptor 4-Mediated Activation of p38 Mitogen-Activated Protein Kinase is a Determinant of Respiratory Virus Entry and Tropism. J. Virol. 2010, 84, 11359–11373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoenfelt, J.; Mitkus, R.J.; Zeisler, R.; Spatz, R.O.; Powell, J.; Fenton, M.J.; Squibb, K.A.; Medvedev, A.E. Involvement of TLR2 and TLR4 in Inflammatory Immune Responses Induced by Fine and Coarse Ambient Air Particulate Matter. J. Leucoc. Biol. 2009, 86, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, R.T.; Carneiro, L.A.M.; Bozza, M.T. Fungal Surface and Innate Immune Recognition of Filamentous Fungi. Front. Microbiol. 2011, 2, 248. [Google Scholar] [CrossRef] [Green Version]
- Bourgeois, C.; Kuchler, K. Fungal Pathogens—a Sweet and Sour Treat for Toll-like Receptors. Front. Cell Infect. Microbiol. 2012, 2, 142. [Google Scholar] [CrossRef] [Green Version]
- Hosoki, K.; Boldogh, I.; Sur, S. Innate Response to Pollen Allergens. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Dahl, Å. Pollen Lipids can Play a Role in Allergic Airway Inflammation. Front. Immunol. 2018, 9, 2816. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Neely, G.; Yaghubian-Malhami, R.; Perkmann, T.; Van Loo, G.; Ermolaeva, M.; Veldhuizen, R.; Leung, Y.C.; Wang, H.; et al. Identification of Oxidative Stress and Toll-like Receptor 4 Signaling as a Key Pathway of Acute Lung Injury. Cell 2008, 133, 235–249. [Google Scholar] [CrossRef]
- Hu, R.; Xu, H.; Jiang, H.; Zhang, Y.; Sun, Y. The Role of TLR4 in the Pathogenesis of Indirect Acute Lung Injury. Front. Biosci. 2013, 18, 1244–1255. [Google Scholar]
- Palti, Y. Toll-like Receptors in Bony Fish: From Genomics to Function. Develop. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar] [CrossRef]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.-M.; Hoffman, J.A. The Dorsoventral Regulatory Gene Cassette spätzle/Toll/cactus Controls the Potent Antifugal Response in Drosophila Adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef] [Green Version]
- Bell, J.; Mullen, G.E.; Leifer, C.A.; Mazzoni, A.; Davies, D.R.; Segal, D.M. Leucine-rich Repeats and Pathogen Recognition in Toll-like Receptors. Trends Immunol. 2003, 24, 528–533. [Google Scholar] [CrossRef]
- Yee, V.C.; Pratt, K.P.; Cote, H.C.; Le Trong, I.; Chung, D.W.; Davie, E.W.; Stenkamp, R.E.; Teller, D.C. Crystal structure of a 30 kDa C-Terminal Fragment from the γ Chain of Human Fibrinogen. Structure 1997, 5, 125–138. [Google Scholar] [CrossRef] [Green Version]
- Doolittle, R.F.; McNamara, K.; Lin, K. Correlating Structure and Function during the Evolution of Fibrinogen-Related Domains. Prot. Sci. 2012, 21, 1808–1823. [Google Scholar] [CrossRef] [Green Version]
- Zuliani-Alvarez, L.; Midwood, K.S. Fibrinogen-Related Proteins in Tissue Repair: How a Unique Domain with a Common Structure Controls Diverse Aspects of Wound Healing. Adv. Wound Care 2015, 4, 273–285. [Google Scholar] [CrossRef] [Green Version]








| Tree Pollens |
| Acer, Alnus, Betula, Carya, Cupressaceae, Cyperaceae, Fagus, Fraxinus, Juglans, Morus, Olea, Quercus, Pinaceae, Platanus, Populus, Salix, Tilia, Ulmus |
| Weed Pollens |
| Ambrosia, Artemisia, Asteraceae (excluding Ambrosia and Artemisia), Chenopodiaceae/Amaranthaceae, Liquidambar, Plantago, Rumex, Typha, Urticaceae |
| Grass Pollen |
| Gramineae/Poaceae |
| Mold Spores |
| Alternaria, Botrytis, Cercospora, Chaetomium, Cladosporium, Coprinus-type, Curvularia, Diatrypaceae, Dreshlera/Helminthosporium, Epicoccum, Fusarium, Ganaderma, Leptosphaeria-type, Nigrospora, Oidium/Erysiphe, Penicillium/Aspergillus, Periconia, Peronospora, Pithomyces, Pleospora, Polythrincium, Rusts, Smuts/Myxomycetes, Stemphylium, Torula, undifferentiated Ascospores, undifferentiated Basidiospores, other fungi |
| Appearance | |||||||
| Year | Date of Assigned Min. | Date of Assigned Max. | ILI0, Presentations (95%) | k, d−1 (95%) | Half-life, d | C, Presentations (95%) | R2 |
| 2014 | 1/9/2015 | 3/30/2015 | 5 (3–7) | 0.033 (0.028–0.042) | 21 | 36 (33–39) | 0.761 |
| 2015 | 10/27/2015 | 2/22/2016 | 1 (1–2) | 0.035 (0.029–0.043) | 20 | 40 (39–42) | 0.699 |
| 2016 | 10/22/2016 | 2/19/2017 | 2 (1–6) | 0.029 (0.022–0.038) | 24 | 35 (28–40) | 0.747 |
| 2017 | 10/9/2017 | 1/28/2018 | 19 (12–22) | 0.021 (0.019–0.025) | 34 | 0 (0–11) | 0.882 |
| 2018 | 10/28/2018 | 3/20/2019 | 39 (36–42) | 0.004 (0.003–0.005) | 185 | 0 (0–0) | 0.346 |
| 2019 | 11/11/2019 | 1/27/2020 | 49 (41–56) | 0.016 (0.014–0.019) | 43 | 0 (0–0) | 0.673 |
| 2020 | - | - | - | - | - | - | - |
| Median | 12 | 0.025 | 29 | 18 | |||
| Disappearance | |||||||
| Year | Date of Assigned Max. | Date of Assigned Min. | ILI0, Presentations (95%) | k, d−1 (95%) | Half-life, d | C, Presentations (95%) | R2 |
| 2014 | - | - | - | - | - | - | - |
| 2015 | 3/30/2015 | 8/4/2015 | 66 (61–72) | 0.041 (0.034–0.049) | 17 | 29 (26–31) | 0.820 |
| 2016 | 2/22/2016 | 8/4/2016 | 113 (107–118) | 0.031 (0.027–0.035) | 22 | 18 (16–21) | 0.899 |
| 2017 | 2/19/2017 | 9/3/2017 | 105 (99–110) | 0.019 (0.017–0.022) | 36 | 13 (9–16) | 0.881 |
| 2018 | 1/28/2018 | 8/3/2018 | 155 (148–161) | 0.028 (0.026–0.031) | 24 | 20 (18–23) | 0.935 |
| 2019 | 3/20/2019 | 8/20/2019 | 63 (59–68) | 0.024 (0.020–0.029) | 29 | 16 (13–19) | 0.839 |
| 2020 | 3/18/2020 | 7/18/2020 | 145 (136–154) | 0.026 (0.021–0.030) | 27 | 10 (1–7) | 0.897 |
| Median | 109 | 0.027 | 26 | 17 | |||
| Bioaerosol | Particle Count | % Total |
|---|---|---|
| Cladosporium | 6,064,682 | 47.287 |
| Undifferentiated Ascospores | 3,598,475 | 28.058 |
| Smuts/Myxomycetes | 820,505 | 6.398 |
| Coprinus-type | 600,774 | 4.684 |
| Ganoderma | 368,568 | 2.874 |
| Alternaria | 224,356 | 1.749 |
| Cercospora | 208,950 | 1.629 |
| Diatrypaceae | 116,333 | 0.907 |
| Penicillium/Aspergillus | 109,272 | 0.852 |
| Epicoccum | 97,962 | 0.764 |
| Undifferentiated Basidiospores | 93,886 | 0.732 |
| Rusts | 93,107 | 0.726 |
| Dreshslera/Helminthosporium | 79,746 | 0.622 |
| Pithomyces | 45,102 | 0.352 |
| Leptosphaeria-type | 40,322 | 0.314 |
| Nigrospora | 36,701 | 0.286 |
| Torula | 32,652 | 0.255 |
| Morus | 28,019 | 0.218 |
| Periconia | 26,389 | 0.206 |
| Chaetomium | 14,408 | 0.112 |
| Curvularia | 14,166 | 0.11 |
| Oidium/Erysiphe | 12,012 | 0.094 |
| Stemphylium | 11,304 | 0.088 |
| Fusarium | 10,277 | 0.08 |
| Polythrincium | 9714 | 0.076 |
| Populus | 8034 | 0.063 |
| Quercus | 6792 | 0.053 |
| Acer | 6576 | 0.051 |
| Ambrosia | 5896 | 0.046 |
| Gramineae/Poaceae | 5399 | 0.042 |
| Peronospora | 4676 | 0.036 |
| Chenopodiaceae/Amaranthaceae | 4637 | 0.036 |
| Cupressaceae | 4395 | 0.034 |
| Betula | 4132 | 0.032 |
| Pinaceae | 2607 | 0.02 |
| Unidentified Fungi | 2366 | 0.018 |
| Urticaceae | 2337 | 0.018 |
| Plantago | 1584 | 0.012 |
| Pleospora | 1473 | 0.011 |
| Salix | 1284 | 0.01 |
| Artemisia | 1019 | 0.008 |
| Fraxinus | 823 | 0.006 |
| Ulmus | 822 | 0.006 |
| Unidentified Pollen | 699 | 0.005 |
| Rumex | 483 | 0.004 |
| Juglans | 386 | 0.003 |
| Alnus | 281 | 0.002 |
| Platanus | 231 | 0.002 |
| Tilia | 170 | 0.001 |
| Carya | 147 | 0.001 |
| Cyperaceae | 128 | 0.001 |
| Fagus | 120 | 0.001 |
| Liquidambar | 85 | 0.001 |
| Typha | 38 | 0 |
| Asteraceae (excl. Ambrosia and Artemisia) | 38 | 0 |
| Olea | 18 | 0 |
| Botrytis | 0 | 0 |
| Disappearance | |||||||
| Year | Date of Assigned Maximum, A | Mold Spore Count on A, particles/m3 | Date of Last Measurement, B | Mold Spore Count on B, particles/m3 | Interval (A to B), d | ||
| 2015 | 9/2/2015 | 76,403 | 10/16/2015 | 10,344 | 44 | ||
| 2016 | 10/6/2016 | 101,028 | 10/21/2016 | 7203 | 15 | ||
| 2017 | 9/25/2017 | 63,929 | 10/20/2017 | 10,519 | 25 | ||
| 2018 | 9/5/2018 | 80,759 | 10/12/2018 | 22,466 | 37 | ||
| 2019 | 9/20/2019 | 43,414 | 9/30/2019 | 12,920 | 10 | ||
| 2020 | 9/4/2020 | 57,100 | 10/13/2020 | 5200 | 39 | ||
| Median | 70,166 | 10,432 | 31 | ||||
| Appearance | |||||||
| Year | Date of First Observed Appearance | Date of Assigned Maximum | Interval, d | M0, particles/m3 (95%) | k, d−1 (95%) | Half-life, d | R2 |
| 2015 | 3/16/2015 | 9/2/2015 | 170 | 7350 (5427–9923) | 0.008 (0.006–0.011) | 84 | 0.211 |
| 2016 | 3/21/2016 | 10/6/2016 | 199 | 3759 (2788–5081) | 0.012 (0.010–0.014) | 59 | 0.478 |
| 2017 | 2/20/2017 | 9/25/2017 | 217 | 2682 (4418–7437) | 0.011 (0.006–0.009) | 65 | 0.395 |
| 2018 | 3/1/2018 | 9/5/2018 | 188 | 2256 (1489–3142) | 0.015 (0.013–0.018) | 45 | 0.572 |
| 2019 | 4/1/2019 | 9/24/2019 | 176 | 3137 (1399–3632) | 0.013 (0.012–0.019) | 52 | 0.634 |
| 2020 | 3/23/2020 | 9/4/2020 | 165 | 4044 (2820–5624) | 0.012 (0.010–0.015) | 56 | 0.479 |
| Median | 188 | 3137 | 0.012 | 59 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, R.B.; Shah, R.D.; Retzinger, D.G.; Retzinger, A.C.; Retzinger, D.A.; Retzinger, G.S. Competing Bioaerosols May Influence the Seasonality of Influenza-Like Illnesses, including COVID-19. The Chicago Experience. Pathogens 2021, 10, 1204. https://doi.org/10.3390/pathogens10091204
Shah RB, Shah RD, Retzinger DG, Retzinger AC, Retzinger DA, Retzinger GS. Competing Bioaerosols May Influence the Seasonality of Influenza-Like Illnesses, including COVID-19. The Chicago Experience. Pathogens. 2021; 10(9):1204. https://doi.org/10.3390/pathogens10091204
Chicago/Turabian StyleShah, Richa B., Rachna D. Shah, Damien G. Retzinger, Andrew C. Retzinger, Deborah A. Retzinger, and Gregory S. Retzinger. 2021. "Competing Bioaerosols May Influence the Seasonality of Influenza-Like Illnesses, including COVID-19. The Chicago Experience" Pathogens 10, no. 9: 1204. https://doi.org/10.3390/pathogens10091204
APA StyleShah, R. B., Shah, R. D., Retzinger, D. G., Retzinger, A. C., Retzinger, D. A., & Retzinger, G. S. (2021). Competing Bioaerosols May Influence the Seasonality of Influenza-Like Illnesses, including COVID-19. The Chicago Experience. Pathogens, 10(9), 1204. https://doi.org/10.3390/pathogens10091204