Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Captures and Blood Sampling
2.3. Kinetoplastid Detection
2.4. Bioinformatics Analysis
2.5. Phylogenetic Analysis
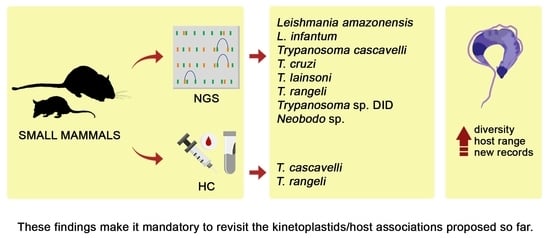
3. Results
| Species | Mdom (3/4) | Tmac (1/1) | Omam (9/12) | Tfos (9/10) | Clat (2/5) | Hcha (1/1) |
|---|---|---|---|---|---|---|
| Leishmania amazonensis (ASV8) | ||||||
| Leishmania infantum (ASV9) | ||||||
| Trypanosoma cascavelli (ASV6) | ||||||
| Trypanosoma cruzi DTU TcI (ASV1) | ||||||
| Trypanosoma cruzi DTU TcI (ASV2) | ||||||
| Trypanosoma cruzi DTU TcII (ASV3) | ||||||
| Trypanosoma lainsoni (ASV7) | ||||||
| Trypanosoma rangeli A (ASV11) | ||||||
| Trypanosoma rangeli B (ASV4) | ||||||
| Trypanosoma rangeli E (ASV12) | ||||||
| Trypanosoma sp. DID (ASV5) |

4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cavalier-Smith, T. Eukaryote kingdoms: Seven or nine? Biosystems 1981, 14, 461–481. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [Green Version]
- Lukeš, J.; Skalický, T.; Týč, J.; Votýpka, J.; Yurchenko, V. Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 2014, 195, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Votýpka, J.; D’Avila-Levy, C.M.; Grellier, P.; Maslov, D.A.; Lukeš, J.; Yurchenko, V. New Approaches to Systematics of Trypanosomatidae: Criteria for Taxonomic (Re)description. Trends Parasitol. 2015, 31, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Belyaev, A.O.; Zagumyonnyi, D.G.; Mylnikov, A.P.; Tikhonenkov, D.V. The Morphology, Ultrastructure and Molecular Phylogeny of a New Soil-Dwelling Kinetoplastid Avlakibodo gracilis gen. et sp. nov. (Neobodonida; Kinetoplastea). Protist 2022, 173, 125885. [Google Scholar] [CrossRef]
- Vickerman, K. The Diversity of the Kinetoplastid Flagellates. In Biology of the Kinetoplastida; Lumsden, W.H.R., Evans, D.A., Eds.; Academic Press: London, UK, 1976; Volume 1, pp. 1–34. [Google Scholar]
- D’Avila-Levy, C.M.; Boucinha, C.; Kostygov, A.; Santos, H.L.C.; Morelli, K.A.; Grybchuk-Ieremenko, A.; Duval, L.; Votypka, J.; Yurchenko, V.; Grellier, P.; et al. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Mem. Inst. Oswaldo Cruz 2015, 110, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Hoare, C.A. The Classification of Mammalian Trypanosomes. In Ergebnisse der Mikrobiologie Immunitätsforschung und Experimentellen Therapie; Springer: Berlin/Heidelberg, Germany, 1966; pp. 43–57. [Google Scholar]
- Kaufer, A.; Ellis, J.; Stark, D.; Barratt, J. The evolution of trypanosomatid taxonomy. Parasites Vectors 2017, 10, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostygov, A.Y.; Frolov, A.O.; Malysheva, M.N.; Ganyukova, A.I.; Chistyakova, L.V.; Tashyreva, D.; Tesařová, M.; Spodareva, V.V.; Režnarová, J.; Macedo, D.H.; et al. Vickermania gen. nov., trypanosomatids that use two joined flagella to resist midgut peristaltic flow within the fly host. BMC Biol. 2020, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Tesařová, M.; Yurchenko, V.; Votýpka, J. Characterization of a new cosmopolitan genus of trypanosomatid parasites, Obscuromonas gen. nov. (Blastocrithidiinae subfam. nov.). Eur. J. Protistol. 2021, 79, 125778. [Google Scholar] [CrossRef] [PubMed]
- Maslov, D.A.; Opperdoes, F.; Kostygov, A.; Hashimi, H.; Lukeš, J.; Yurchenko, V. Recent advances in trypanosomatid research: Genome organization, expression, metabolism, taxonomy and evolution. Parasitology 2019, 146, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.G.; Stevens, J.R.; Lukeš, J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006, 22, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Herrera, H.; Rocha, F.; Lisboa, C.; Rademaker, V.; Mourão, G.; Jansen, A.M. Food web connections and the transmission cycles of Trypanosoma cruzi and Trypanosoma evansi (Kinetoplastida, Trypanosomatidae) in the Pantanal Region, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 380–387. [Google Scholar] [CrossRef]
- Herrera, H.; Rademaker, V.; Abreu, U.; D’Andrea, P.; Jansen, A. Variables that modulate the spatial distribution of Trypanosoma cruzi and Trypanosoma evansi in the Brazilian Pantanal. Acta Trop. 2007, 102, 55–62. [Google Scholar] [CrossRef]
- Herrera, H.; Dávila, A.; Norek, A.; Abreu, U.; Souza, S.; D’Andrea, P.; Jansen, A. Enzootiology of Trypanosoma evansi in Pantanal, Brazil. Vet. -Parasitol. 2004, 125, 263–275. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Porfirio, G.E.; Santos, F.M.; de Macedo, G.C.; Barreto, W.T.G.; Campos, J.B.V.; Meyers, A.C.; André, M.R.; Perles, L.; de Oliveira, C.E.; das Chagas Xavier, S.C.; et al. Maintenance of Trypanosoma cruzi, T. evansi and Leishmania spp. by domestic dogs and wild mammals in a rural settlement in Brazil-Bolivian border. Int. J. Parasitol. Parasites Wildl. 2018, 7, 398–404. [Google Scholar] [CrossRef]
- Rademaker, V.; Herrera, H.; Raffel, T.; D’Andrea, P.; Freitas, T.; Abreu, U.; Hudson, P.; Jansen, A. What is the role of small rodents in the transmission cycle of Trypanosoma cruzi and Trypanosoma evansi (Kinetoplastida Trypanosomatidae)? A study case in the Brazilian Pantanal. Acta Trop. 2009, 111, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.M.; Jansen, A.M.; Mourão, G.D.M.; Jurberg, J.; Nunes, A.P.; Herrera, H. Triatominae (Hemiptera, Reduviidae) in the Pantanal region: Association with Trypanosoma cruzi, different habitats and vertebrate hosts. Rev. Soc. Bras. Med. Trop. 2015, 48, 532–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, F.M.; Barreto, W.T.G.; de Macedo, G.C.; Barros, J.H.D.S.; Xavier, S.C.D.C.; Garcia, C.M.; Mourão, G.; de Oliveira, J.; Rimoldi, A.R.; Porfírio, G.E.D.O.; et al. The reservoir system for Trypanosoma (Kinetoplastida, Trypanosomatidae) species in large neotropical wetland. Acta Trop. 2019, 199, 105098. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.D.; Gofton, A.; Paparini, A.; Codello, A.; Greay, T.; Gillett, A.; Warren, K.; Irwin, P.; Ryan, U. Increased genetic diversity and prevalence of co-infection with Trypanosoma spp. in koalas (Phascolarctos cinereus) and their ticks identified using next-generation sequencing (NGS). PLoS ONE 2017, 12, e0181279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, M.S.; Lima, L.; Xavier, S.C.D.C.; Herrera, H.M.; Rocha, F.L.; Roque, A.L.R.; Teixeira, M.M.G.; Jansen, A.M. Uncovering Trypanosoma spp. diversity of wild mammals by the use of DNA from blood clots. Int. J. Parasitol. Parasites Wildl. 2019, 8, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Carr, I.M.; Robinson, J.; Dimitriou, R.; Markham, A.F.; Morgan, A.; Bonthron, D. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics 2009, 25, 3244–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantin, Y.S.; Neverov, A.D.; Favorov, A.V.; Alvarez-Figueroa, M.V.; Braslavskaya, S.I.; Gordukova, M.A.; Karandashova, I.V.; Kuleshov, K.V.; Myznikova, A.I.; Polishchuk, M.S.; et al. Base-Calling Algorithm with Vocabulary (BCV) Method for Analyzing Population Sequencing Chromatograms. PLoS ONE 2013, 8, e54835. [Google Scholar] [CrossRef]
- Paparini, A.; Jackson, B.; Ward, S.; Young, S.; Ryan, U.M. Multiple Cryptosporidium genotypes detected in wild black rats (Rattus rattus) from northern Australia. Exp. Parasitol. 2012, 131, 404–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, A.; Austen, J.; Gillett, A.; Warren, K.; Paparini, A.; Irwin, P.; Ryan, U. First report of Trypanosoma vegrandis in koalas (Phascolarctos cinereus). Parasitol. Int. 2016, 65, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Botero, A.; Thompson, C.K.; Peacock, C.S.; Clode, P.L.; Nicholls, P.K.; Wayne, A.F.; Lymbery, A.J.; Thompson, R.A. Trypanosomes genetic diversity, polyparasitism and the population decline of the critically endangered Australian marsupial, the brush tailed bettong or woylie (Bettongia penicillata). Int. J. Parasitol. Parasites Wildl. 2013, 2, 77–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McINNES, L.M.; Gillett, A.; Hanger, J.; Reid, S.A.; Ryan, U.M. The potential impact of native Australian trypanosome infections on the health of koalas (Phascolarctos cinereus). Parasitology 2011, 138, 873–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dario, M.A.; Furtado, C.; Lisboa, C.V.; de Oliveira, F.; Santos, F.M.; D’Andrea, P.S.; Roque, A.L.R.; Xavier, S.C.D.C.; Jansen, A.M. Trypanosomatid Richness Among Rats, Opossums, and Dogs in the Caatinga Biome, Northeast Brazil, a Former Endemic Area of Chagas Disease. Front. Cell. Infect. Microbiol. 2022, 12, 851903. [Google Scholar] [CrossRef]
- Dario, M.A.; Moratelli, R.; Schwabl, P.; Jansen, A.M.; Llewellyn, M.S. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Negl. Trop. Dis. 2017, 11, e0005790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.D.S.V.; Abdon, M.D.M. Delimitation of the Brazilian Pantanal and Its Subregions. Pesqui. Agropecuária Bras. 1998, 33, 1703–1711. [Google Scholar]
- Sano, N.Y.; Herrera, H.M.; Porfirio, G.E.D.O.; Santos, F.M. Understory use by terrestrial small mammals in an unflooded forest patch in the Pantanal floodplain. Mammalia 2021, 85, 164–167. [Google Scholar] [CrossRef]
- Antunes, P.C.; Oliveira-Santos, L.G.R.; Tomas, W.M.; Forester, J.D.; Fernandez, F.A.S. Disentangling the effects of habitat, food, and intraspecific competition on resource selection by the spiny rat, Thrichomys fosteri. J. Mammal. 2016, 97, 1738–1744. [Google Scholar] [CrossRef]
- Vallejo, G.A.; Guhl, F.; Chiari, E.; Macedo, A.M. Species Specific Detection of Trypanosoma cruzi and Trypanosoma rangeli in Vector and Mammalian Hosts by Polymerase Chain Reaction Amplification of Kinetoplast Minicircle DNA. Acta Trop. 1999, 72, 203–212. [Google Scholar] [CrossRef]
- Brandão, E.M.V.; Xavier, S.C.C.; Rocha, F.L.; Lima, C.F.M.; Candeias, Z.; Lemos, F.G.; Azevedo, F.C.; Jansen, A.M.; Roque, A.L.R. Wild and Domestic Canids and Their Interactions in the Transmission Cycles of Trypanosoma Cruzi and Leishmania spp. in an Area of the Brazilian Cerrado. Pathogens 2020, 9, 818. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Clark, P.; Averis, S.; Lymbery, A.J.; Wayne, A.F.; Morris, K.D.; Thompson, R.C.A. Trypanosomes in a declining species of threatened Australian marsupial, the brush-tailed bettong Bettongia penicillate (Marsupialia: Potoroidae). Parasitology 2008, 135, 1329–1335. [Google Scholar] [CrossRef] [Green Version]
- The R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Polzin, T.; Daneshmand, S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003, 31, 12–20. [Google Scholar] [CrossRef]
- Hamilton, P.B.; Gibson, W.C.; Stevens, J.R. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phylogenet. Evol. 2007, 44, 15–25. [Google Scholar] [CrossRef]
- Dario, M.A.; Lisboa, C.V.; Xavier, S.C.D.C.; D’Andrea, P.S.; Roque, A.L.R.; Jansen, A.M. Trypanosoma Species in Small Nonflying Mammals in an Area with a Single Previous Chagas Disease Case. Front. Cell. Infect. Microbiol. 2022, 12, 812708. [Google Scholar] [CrossRef]
- Nantes, W.A.G.; Santos, F.M.; De Macedo, G.C.; Barreto, W.T.G.; Gonçalves, L.R.; Rodrigues, M.S.; Chulli, J.V.M.; Rucco, A.C.; Assis, W.D.O.; Porfírio, G.E.D.O.; et al. Trypanosomatid species in Didelphis albiventris from urban forest fragments. Parasitol. Res. 2021, 120, 223–231. [Google Scholar] [CrossRef]
- Dario, M.A.; Lisboa, C.V.; Costa, L.M.; Moratelli, R.; Nascimento, M.P.; Costa, L.P.; Leite, Y.L.R.; Llewellyn, M.S.; Xavier, S.C.D.C.; Roque, A.L.R.; et al. High Trypanosoma spp. diversity is maintained by bats and triatomines in Espírito Santo state, Brazil. PLoS ONE 2017, 12, e0188412. [Google Scholar] [CrossRef]
- Viola, L.B.; Campaner, M.; Takata, C.S.A.; Ferreira, R.C.; Rodrigues, A.C.; Freitas, R.A.; Duarte, M.R.; Grego, K.F.; Barrett, T.V.; Camargo, E.P.; et al. Phylogeny of snake trypanosomes inferred by SSU rDNA sequences, their possible transmission by phlebotomines, and taxonomic appraisal by molecular, cross-infection and morphological analysis. Parasitology 2008, 135, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Barrios, S.P.G.; Pereira, L.E.; Monaco, N.Z.N.; Graciolli, G.; Casaril, A.E.; Infran, J.D.O.M.; de Oliveira, E.F.; Fernandes, W.D.S.; Filho, A.C.P.; De Oliveira, A.G. Synanthropy and diversity of Phlebotominae in an area of intense transmission of visceral leishmaniasis in the South Pantanal floodplain, Midwest Brazil. PLoS ONE 2019, 14, e0215741. [Google Scholar] [CrossRef] [PubMed]
- Braga-Miranda, L.C.; Miranda, M.; Galati, E.A.B. Phlebotomine fauna in a rural area of the Brazilian Pantanal. Rev. Saude Publica 2006, 40, 324–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, E.F.; Oshiro, E.T.; Fernandes, W.S.; Ferreira, A.M.T.; de Oliveira, A.G.; Galati, E.A.B. Vector Competence of Lutzomyia cruzi Naturally Demonstrated for Leishmania infantum and Suspected for Leishmania amazonensis. Am. J. Trop. Med. Hyg. 2017, 96, 178–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, E.D.C.; Pereira, A.A.S.; Silveira, M.; Margonari, C.; Marcon, G.E.B.; França, A.D.O.; Castro, L.S.; Bordignon, M.O.; Fischer, E.; Tomas, W.M.; et al. Leishmania (V.) braziliensis infecting bats from Pantanal wetland, Brazil: First records for Platyrrhinus lineatus and Artibeus planirostris. Acta Trop. 2017, 172, 217–222. [Google Scholar] [CrossRef]
- Barrios, S.P.G.; Pereira, L.E.; Casaril, A.E.; Infran, J.D.O.M.; Fernandes, W.D.S.; Oshiro, E.T.; Galati, E.A.B.; Graciolli, G.; Filho, A.C.P.; De Oliveira, A.G. Phlebotominae (Diptera: Psychodidae) and Biomes in the State of Mato Grosso do Sul, Brazil. J. Med. Èntomol. 2020, 57, 1882–1904. [Google Scholar] [CrossRef]
- DOS Santos, S.O.; Arias, J.; Ribeiro, A.A.; Hoffmann, M.D.P.; DE Freitas, R.A.; Malacco, M.A.F. Incrimination of Lutzomyia cruzi as a vector of American Visceral Leishmaniasis. Med. Vet.-Èntomol. 1998, 12, 315–317. [Google Scholar] [CrossRef]
- De Oliveira, E.F.; Oshiro, E.T.; Fernandes, W.D.S.; Murat, P.G.; De Medeiros, M.J.; Souza, A.; De Oliveira, A.G.; Galati, E.A.B. Experimental infection and transmission of Leishmania by Lutzomyia cruzi (Diptera: Psychodidae): Aspects of the ecology of parasite-vector interactions. PLoS Negl. Trop. Dis. 2017, 11, e0005401. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Castañeda, A.; Patiño, L.H.; Muñoz, M.; Ayala, M.S.; Segura, M.; Bautista, J.; Shaban, M.V.; Paniz-Mondolfi, A.; Ramírez, J.D. Amplicon-based next-generation sequencing reveals the co-existence of multiple Leishmania species in patients with visceral leishmaniasis. Int. J. Infect. Dis. 2022, 115, 35–38. [Google Scholar] [CrossRef]
- da Silva, F.M.; Marcili, A.; Lima, L.; Cavazzana, M.; Ortiz, P.; Campaner, M.; Takeda, G.; Paiva, F.; Nunes, V.; Camargo, E.; et al. Trypanosoma rangeli isolates of bats from Central Brazil: Genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop. 2009, 109, 199–207. [Google Scholar] [CrossRef]
- DA Silva, F.M.; Rodrigues, A.C.; Campaner, M.; Takata, C.S.A.; Brigido, M.C.; Junqueira, A.C.V.; Coura, J.R.; Takeda, G.F.; Shaw, J.J.; Teixeira, M.M.G. Randomly amplified polymorphic DNA analysis of Trypanosoma rangeli and allied species from human, monkeys and other sylvatic mammals of the Brazilian Amazon disclosed a new group and a species-specific marker. Parasitology 2004, 128, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Dario, M.; Pavan, M.; Rodrigues, M.; Lisboa, C.; Kluyber, D.; Desbiez, A.; Herrera, H.; Roque, A.; Lima, L.; Teixeira, M.; et al. Trypanosoma rangeli Genetic, Mammalian Hosts, and Geographical Diversity from Five Brazilian Biomes. Pathogens 2021, 10, 736. [Google Scholar] [CrossRef] [PubMed]
- de Lima, J.S.; Rocha, F.L.; Alves, F.M.; Lorosa, E.S.; Jansen, A.M.; Mourão, G.D.M. Infestation of arboreal nests of coatis by triatomine species, vectors of Trypanosoma cruzi, in a large Neotropical wetland. J. Vector Ecol. 2015, 40, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Flegontova, O.; Flegontov, P.; Londoño, P.A.C.; Walczowski, W.; Šantić, D.; Edgcomb, V.P.; Lukeš, J.; Horák, A. Environmental determinants of the distribution of planktonic diplonemids and kinetoplastids in the oceans. Environ. Microbiol. 2020, 22, 4014–4031. [Google Scholar] [CrossRef]
- Szőke, K.; Sándor, A.D.; Boldogh, S.A.; Görföl, T.; Votýpka, J.; Takács, N.; Estók, P.; Kováts, D.; Corduneanu, A.; Molnár, V.; et al. DNA of free-living bodonids (Euglenozoa: Kinetoplastea) in bat ectoparasites: Potential relevance to the evolution of parasitic trypanosomatids. Acta Vet.-Hung. 2017, 65, 531–540. [Google Scholar] [CrossRef] [Green Version]
- Votýpka, J.; Petrželková, K.J.; Brzoňová, J.; Jirků, M.; Modrý, D.; Lukeš, J. How monoxenous trypanosomatids revealed hidden feeding habits of their tsetse fly hosts. Folia Parasitol. 2021, 68, 019. [Google Scholar] [CrossRef]
- Spisák, S.; Solymosi, N.; Ittzés, P.; Bodor, A.; Kondor, D.; Vattay, G.; Barták, B.K.; Sipos, F.; Galamb, O.; Tulassay, Z.; et al. Complete Genes May Pass from Food to Human Blood. PLoS ONE 2013, 8, e69805. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, R.D.C.; Campos, R.C.; Xavier-Filho, N.L.; Olifiers, N.; Gompper, M.E.; Mourão, G. Intraspecific, interspecific, and seasonal differences in the diet of three mid-sized carnivores in a large neotropical wetland. Acta Thériol. 2014, 59, 13–23. [Google Scholar] [CrossRef]
- Reimann, M.M.; Torres-Santos, E.C.; de Souza, C.S.F.; Andrade-Neto, V.V.; Jansen, A.M.; Brazil, R.P.; Roque, A.L.R. Oral and Intragastric: New Routes of Infection by Leishmania braziliensis and Leishmania infantum? Pathogens 2022, 11, 688. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, F.M.; Sano, N.Y.; Liberal, S.C.; Dario, M.A.; Nantes, W.A.G.; Alves, F.M.; da Silva, A.R.; De Oliveira, C.E.; Roque, A.L.R.; Herrera, H.M.; et al. Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome. Pathogens 2022, 11, 1205. https://doi.org/10.3390/pathogens11101205
Santos FM, Sano NY, Liberal SC, Dario MA, Nantes WAG, Alves FM, da Silva AR, De Oliveira CE, Roque ALR, Herrera HM, et al. Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome. Pathogens. 2022; 11(10):1205. https://doi.org/10.3390/pathogens11101205
Chicago/Turabian StyleSantos, Filipe Martins, Nayara Yoshie Sano, Sany Caroline Liberal, Maria Augusta Dario, Wesley Arruda Gimenes Nantes, Fernanda Moreira Alves, Alanderson Rodrigues da Silva, Carina Elisei De Oliveira, André Luiz Rodrigues Roque, Heitor Miraglia Herrera, and et al. 2022. "Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome" Pathogens 11, no. 10: 1205. https://doi.org/10.3390/pathogens11101205
APA StyleSantos, F. M., Sano, N. Y., Liberal, S. C., Dario, M. A., Nantes, W. A. G., Alves, F. M., da Silva, A. R., De Oliveira, C. E., Roque, A. L. R., Herrera, H. M., & Jansen, A. M. (2022). Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome. Pathogens, 11(10), 1205. https://doi.org/10.3390/pathogens11101205