Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets
Abstract
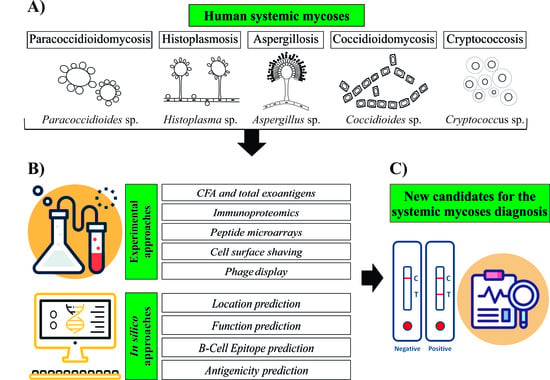
:1. Human Systemic Mycosis
2. Neglected Human Systemic Mycoses Diagnosis
2.1. Paracoccidioidomycosis
| Mycosis | Diagnostic Test | Human Specimen | Time until Results | Accuracy | Advantage | Disadvantage | Infrastructural Resources | References |
|---|---|---|---|---|---|---|---|---|
| Paracoccidioidomycosis (PCM) | Double Immunodiffusion test | Serum | This technique requires much time. | Specificity 100% and Sensitivity 65–100% | The choice method in patients with suspected PCM without false positive results or cross reaction. | Low accuracy for determination of the patient’s cellular immunity important during the therapy. | Laborious. However, it is not expensive. | [12,13,17] |
| ELISA | Serum | This technique is fast. | Specificity 88–95%. Sensitivity 100% | Fast and suitable for PCM high-throughput screening. | Cross-reaction with Histoplasmmosis | Requires specific equipment and automatization. | [15,16] | |
| Nucleic Acid testing | Blood, and clinical specimen | Result is obtained in a few hours. | Specificity 100% | Genotypic studies and clinical diagnosis performed directly from samples. | Need to standardize techniques based on DNA amplification for its real implementation. | Requires a specific and high-cost equipment. | [21] | |
| Histoplasmosis | Immunodiffusion | Serum | This technique requires much time. | Sensitivity 60–70% | Rapid turnaround time | Not be used in immunocompromised individuals, since this group may present increases in false-negative results due to the compromise of the humoral response. | Low costs and simple infrastructure | [22] |
| Complement Fixation | Serum | This technique requires much time. | Sensitivity 60–70% | Rapid turnaround time | Laborious technique and requires well trained personnel | [22] | ||
| Enzyme Immunoassay | Serum, Plasma, Urine/CSF/BAL/Other Body Fluid. | Fast. (approximately 1.5 hours) | Sensitivity ranges 95–100% in urine, over 90% in serum and BAL antigens and 78% in cerebral spinal fluid (CSF) | Particularly important in AIDS patients who have disseminated histoplasmosis and who have large fungal burden | Serologic cross-reactions to Histoplasma-like antigens with Blastomycosis, Coccidioidomycosis, PCM and Aspergillosis | Requirement of specialized laboratories, expensive equipment, and well-trained personnel | [22,23] | |
| Nucleic Acid testing | Blood and other body fluid. | The results are obtained in a few hours. | Specificity 100% and a sensitivity 67% to 100%. | Genotypic studies and clinical diagnosis performed directly from samples | Need to standardize techniques based on DNA amplification for its real implementation. | Requires specific and high-cost equipment. | [24,25] | |
| Aspergillosis | ELISA assays galactomannan (GM) detection * | Serum, lung transplant recipients, sputum or bronchoalveolar lavage. | This technique is fast. | Sensitivity 60 to 100% and specificity 85 to 98% | GM levels are proportional to fungal burden in tissue, and present prognostic value | Both false positive and false negative results have been reported and cross reactivity. | Performed without the need for specialized equipment and reagents | [26,27,28] |
| Nucleic Acid testing | Serum, Lung transplant recipients, sputum or bronchoalveolar lavage. | The results are obtained in a few hours. | Specificity 100% | More sensitive and quick diagnosis | The lack of sensitivity and the difficulty in distinguishing between infection and colonization. | Requires specific equipment and has a high cost. | [29,30,31]. | |
| Coccidioidomycosis | Direct examination | Sputum or bronchoalveolar lavage or other biopsy material | This technique is fast. | N/A | The gold standard diagnostic method | The mold form of Coccidioides produces highly infectious arthroconidia as soon as 72 hours after initial growth. | Requires well trained personnel | [30,32] |
| Culture | Sputum or bronchoalveolar lavage or other biopsy material | Requires a lot of time. | N/A | The gold standard diagnostic method | This form represents a significant risk of inhalational exposure to laboratory personnel. | The potential exposure risks associated with aerosolization | [30,32,33] | |
| Enzyme immunoassays | Serum, urinary, and cerebrospinal fluid | This technique is fast. | Sensitivity 88%. Specificity 90% | Antibody detection EIA is a sensitive and specific test, including high-risk patients’ samples, in detection of IgG and IgM antibodies | Maybe insensitive to early infection. | Performed without the need for specialized equipment and reagents | [30,32,34] | |
| Cryptococcosis | Direct examination | Biopsy material | This technique is fast. | Sensitivity 60–90% | More sensitive and quick diagnosis | Lower sensitivity in HIV-negative patients in association with a low fungal burden. | Low-resource method | [35,36] |
| Culture exam | cerebrospinal fluid | 1 to 2 weeks for definitive results | Sensitivity 85–95% | More sensitive. A gold standard for diagnostic | Need longer incubation periods up to three weeks. | The cultures are easily performed in any microbiology laboratory. | [37] | |
| Nucleic Acid testing | Plasma or cerebrospinal fluid | The results are obtained in a few hours. | Specificity 100% | Allows the determination of the Cryptococcus species | Need to standardize techniques based on DNA amplification for its real implementation. | Requires a specific and high-cost equipment. | [38] | |
| lateral flow assay | Plasma or cerebrospinal fluid | This technique is fast. | Sensitivity 90–100% | Provides a rapid diagnosis of cryptococcosis by detecting capsular antigen of Cryptococcus spp. In serum, plasma or CSF. | Low specificity (false positive 11% to 14%) | Low-costs | [39,40] |
2.2. Histoplasmosis
2.3. Coccidioidomycosis
2.4. Aspergillosis
2.5. Cryptococcosis
3. Approaches Used to Search for New Diagnostic Candidates
3.1. Experimental Approaches
3.1.1. Cell-Free Antigens and Total Exoantigens
| Experimental Approaches | Advantage | Disadvantage | Infrastructure | References |
|---|---|---|---|---|
| Cell-free antigens and total exoantigens | Can be used for various purposes, such as immunological diagnostic techniques and to identify new antigenic targets and studies of cellular immunization processes. | The sensitivity and specificity of the tests are related to the production of the antigen, have high possibility of cross-reaction and difficulty in diagnosis at the beginning of the disease. | Low-cost technique and simple to perform. Requires laboratory infrastructure for incubation of microorganism and purification of secreted molecules. | [124,129,130,137,138,139] |
| Immunoproteomics | Has been used successfully for the identification and characterization of antigens applied as new markers for molecular diagnostics, as well as possible candidates for vaccine production used in therapies. | This technique requires antibodies with high selectivity or sensibility and capture of specific antigens in crude samples | Requires sophisticated laboratory infrastructure, trained professionals, and high cost. | [43,140,141,142] |
| Peptide microarrays | Provides an extremely rapid and robust method which allows thousands of targets to be tested simultaneously. | This approach is not sufficient to cover complete proteomes. Furthermore, it is difficult to bind antibodies that need specific conformations and longer sequences. | It is not a high-cost technique, but it requires infrastructure and specialized professionals. | [143,144] |
| Cell surface shaving | Effective in identifying antigenic proteins exposed on the cell surface, which proves to be one of the best targets for host immunity. | This technique is less used in Gram-negative microorganisms, due to the thinner cell wall that does not resist digestion without lysis. | High-cost technique, which requires specialized laboratory and trained professionals. | [145,146] |
| Phage display | Allows rapid identification and isolation of highly specific phage. | Need of phage display libraries construction with stability, quality, and diversity of antibody. Furthermore, difficult to select antibodies against the antigens which are expressed on the surface of rare cells. | Low-cost technique and simple to perform. | [147,148,149] |
| Bioinformatics analysis | It is possible to map a specific antigen and to identify the epitope with great potential for targets in vaccine and diagnosis development. In silico analyses are faster and more cost-effective, and the possibility of identifying proteins that are not expressed in vitro. | The target identified by in silico analysis need experimental confirmation. | Low-resource method. The infrastructure consists of a computer, internet and trained personnel. | [150,151] |
3.1.2. Immunoproteomics
3.1.3. Peptide Microarrays
3.1.4. Cell Surface Shaving
3.1.5. Phage Display
3.2. In Silico Approaches for Antigen Prediction
3.2.1. B-Cell Epitope Prediction
3.2.2. Antigenicity Prediction
3.2.3. Location Prediction
3.2.4. Functional Characterization of Protein
4. Strategies to Improve the Diagnosis of Human Systemic Mycoses Using the Available Technological Approaches
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salzer, H.J.F.; Burchard, G.; Cornely, O.A.; Lange, C.; Rolling, T.; Schmiedel, S.; Libman, M.; Capone, D.; Le, T.; Dalcolmo, M.P.; et al. Diagnosis and Management of Systemic Endemic Mycoses Causing Pulmonary Disease. Respiration 2018, 96, 283–301. [Google Scholar] [CrossRef]
- Yeo, S.F.; Wong, B. Current Status of Nonculture Methods for Diagnosis of Invasive Fungal Infections. Clin. Microbiol. Rev. 2002, 15, 465–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Fahle, G.A.; Kovacs, J.A. Inability to Culture Pneumocystis jirovecii. mBio 2018, 9, e00939-18. [Google Scholar] [CrossRef] [Green Version]
- Martinez, R. New Trends in Paracoccidioidomycosis Epidemiology. J. Fungi 2017, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Bocca, A.L.; Amaral, A.C.; Teixeira, M.M.; Sato, P.K.; Sato, P.; Shikanai-Yasuda, M.A.; Soares Felipe, M.S. Paracoccidioidomycosis: Eco-Epidemiology, Taxonomy and Clinical and Therapeutic Issues. Future Microbiol. 2013, 8, 1177–1191. [Google Scholar] [CrossRef]
- Shikanai-Yasuda, M.A.; Mendes, R.P.; Colombo, A.L.; Queiroz-Telles, F.D.; Kono, A.S.G.; Paniago, A.M.M.; Nathan, A.; do Valle, A.C.F.; Bagagli, E.; Benard, G.; et al. Brazilian Guidelines for the Clinical Management of Paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017, 50, 715–740. [Google Scholar] [CrossRef]
- Shankar, J.; Restrepo, A.; Clemons, K.V.; Stevens, D. A Hormones and the Resistance of Women to Paracoccidioidomycosis. Clin. Microbiol. Rev. 2011, 24, 296–313. [Google Scholar] [CrossRef] [Green Version]
- Terçarioli, G.R.; Bagagli, E.; Reis, G.M.; Theodoro, R.C.; Bosco, S.D.M.G.; Macoris, S.A.D.G.; Richini-Pereira, V.B. Ecological Study of Paracoccidioides brasiliensis in Soil: Growth Ability, Conidia Production and Molecular Detection. BMC Microbiol. 2007, 7, 92. [Google Scholar] [CrossRef] [Green Version]
- Brummer, E.; Castaneda, E.; Restrepo, A. Paracoccidioidomycosis: An Update. Clin. Microbiol. Rev. 1993, 6, 89–117. [Google Scholar] [CrossRef]
- Mendes, R.P.; Cavalcante, R.D.S.; Marques, S.A.; Marques, M.E.A.; Venturini, J.; Sylvestre, T.F.; Paniago, A.; Pereira, A.C.; Silva, J.; Fabro, A.T.; et al. Paracoccidioidomycosis: Current Perspectives from Brazil. Open Microbiol. J. 2017, 11, 224–282. [Google Scholar] [CrossRef] [Green Version]
- Paracoccidioidomycosis, 46th ed.; Franco, M.; da Silva Lacaz, C.; Restrepo-Moreno, A.; Del Negro, G. (Eds.) CRC Press: Boca Raton, FL, USA, 2018; ISBN 9781351075329. [Google Scholar]
- Cano, L.E.; Restrepo, A. Predictive Value of Serologic Tests in the Diagnosis and Follow-up of Patients with Paracoccidioidomycosis. Rev. Inst. De Med. Trop. São Paulo 1987, 29, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreto, T.C.; Marques, M.E.A.; de Oliveira, M.L.S.C.; Moris, D.V.; de Carvalho, L.R.; Mendes, R.P. Accuracy of Routine Diagnostic Tests Used in Paracoccidioidomycosis Patients at a University Hospital. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Perenha-Viana, M.C.Z.; Gonzales, I.A.A.; Brockelt, S.R.; Machado, L.N.C.; Svidzinski, T.I.E. Serological Diagnosis of Paracoccidioidomycosis through a Western Blot Technique. Clin. Vaccine Immunol. 2012, 19, 616–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puccia, R.; Travassos, L.R. The 43-KDa Glycoprotein from the Human Pathogen Paracoccidioides brasiliensis and Its Deglycosylated Form: Excretion and Susceptibility to Proteolysis. Arch. Biochem. Biophys. 1991, 289, 298–302. [Google Scholar] [CrossRef]
- de Fatimada Silva, J.; de Oliveira, H.C.; Marcos, C.M.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Advances and Challenges in Paracoccidioidomycosis Serology Caused by Paracoccidioides Species Complex: An Update. Diagn. Microbiol. Infect. Dis. 2016, 84, 87–94. [Google Scholar] [CrossRef]
- Soares Mendes Giannini, M.J.; Bueno, J.P.; Aparecida Shikanai-Yasuda, M.; Stolf, A.M.S.; Masuda, A.; Amato Neto, V.; Ferreira, A.W. Antibody Response to the 43 KDa Glycoprotein of Paracoccidioides brasiliensis as a Marker for the Evaluation of Patients under Treatment. Am. J. Trop. Med. Hyg. 1990, 43, 200–206. [Google Scholar] [CrossRef]
- Marques da Silva, S.H.; Queiroz-Telles, F.; Colombo, A.L.; Blotta, M.H.S.L.; Lopes, J.D.; Pires de Camargo, Z. Monitoring Gp43 Antigenemia in Paracoccidioidomycosis Patients during Therapy. J. Clin. Microbiol. 2004, 42, 2419–2424. [Google Scholar] [CrossRef] [Green Version]
- Gegembauer, G.; Araujo, L.M.; Pereira, E.F.; Rodrigues, A.M.; Paniago, A.M.M.; Hahn, R.C.; Camargo, Z.P. de Serology of Paracoccidioidomycosis Due to Paracoccidioides lutzii. PLoS Negl. Trop. Dis. 2014, 8, e2986. [Google Scholar] [CrossRef]
- De Camargo, Z.P. Serology of Paracoccidioidomycosis. Mycopathologia 2008, 165, 289–302. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, B.G.; Hahn, R.C.; de Camargo, Z.P.; Rodrigues, A.M. Molecular Tools for Detection and Identification of Paracoccidioides Species: Current Status and Future Perspectives. J. Fungi 2020, 6, 293. [Google Scholar] [CrossRef]
- Myint, T.; Leedy, N.; Cari, E.V.; Wheat, L.J. Hiv-Associated Histoplasmosis: Current Perspectives. HIV/AIDS-Res. Palliat. Care 2020, 12, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, W.; Myint, T.; Larue, R.; Minderman, M.; Gunn, S.; Wheat, L.J.; Hage, C.A. Diagnosis of Histoplasmosis Using the MVista Histoplasma Galactomannan Antigen Qualitative Lateral Flow-Based Immunoassay: A Multicenter Study. Open Forum Infect. Dis. 2021, 8, ofab454. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, I.C.D.S.; Lana, D.F.D.; Pasqualotto, A.C. The Role of Molecular Tests in the Diagnosis of Disseminated Histoplasmosis. J. Fungi 2020, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Azar, M.M.; Hage, C.A. Laboratory Diagnostics for Histoplasmosis. J. Clin. Microbiol. 2017, 55, 1612–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latge, J.P.; Kobayashi, H.; Debeaupuis, J.P.; Diaquin, M.; Sarfati, J.; Wieruszeski, J.M.; Parra, E.; Bouchara, J.P.; Fournet, B. Chemical and Immunological Characterization of the Extracellular Galactomannan of Aspergillus fumigatus. Infect. Immun. 1994, 62, 5424–5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdaguer, V.; Walsh, T.J.; Hope, W.; Cortez, K.J. Galactomannan Antigen Detection in the Diagnosis of Invasive Aspergillosis. Expert Rev. Mol. Diagn. 2007, 7, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Herrera, S.; Husain, S. Current State of the Diagnosis of Invasive Pulmonary Aspergillosis in Lung Transplantation. Front. Microbiol. 2019, 10, 3273. [Google Scholar] [CrossRef]
- Egger, M.; Jenks, J.D.; Hoenigl, M.; Prattes, J. Blood Aspergillus PCR: The Good, the Bad, and the Ugly. J. Fungi 2020, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Hage, C.A.; Carmona, E.M.; Epelbaum, O.; Evans, S.E.; Gabe, L.M.; Haydour, Q.; Knox, K.S.; Kolls, J.K.; Hassan, M. Erratum: Microbiological Laboratory Testing in the Diagnosis of Fungal Infections in Pulmonary and Critical Care Practice. An Official American Thoracic Society Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2019, 200, 1326. [Google Scholar] [CrossRef]
- Barton, R.C. Laboratory Diagnosis of Invasive Aspergillosis: From Diagnosis to Prediction of Outcome. Scientifica 2013, 2013, 459405. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.; Lainhart, W. A Review of Diagnostics for Coccidiomycosis. Clin. Microbiol. Newsl. 2021, 43, 135–141. [Google Scholar] [CrossRef]
- Twarog, M.; Thompson, G.R. Coccidioidomycosis: Recent Updates. Semin. Respir. Crit. Care Med. 2015, 36, 746–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malo, J.; Holbrook, E.; Zangeneh, T.; Strawter, C.; Oren, E.; Robey, I.; Erickson, H.; Chahal, R.; Durkin, M.; Thompson, C.; et al. Enhanced Antibody Detection and Diagnosis of Coccidioidomycosis with the Miravista IgG and IgM Detection Enzyme Immunoassay. J. Clin. Microbiol. 2017, 55, 893–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulware, D.; Rolfes, M.A.; Rajasingham, R.; Von Hohenberg, M.; Qin, Z.; Taseera, K.; Schutz, C.; Kwizera, R.; Butler, E.K.; Meintjes, G.; et al. Multisite Validation of Cryptococcal Antigen Lateral Flow Assay and Quantification by Laser Thermal Contrast. Emerg. Infect. Dis. 2014, 20, 45. [Google Scholar] [CrossRef]
- Chammard, T.B.; Temfack, E.; Lortholary, O.; Alanio, A. Diagnostic and Therapeutic Strategies in Cryptococcosis: Impact on Outcome. Mem. Inst. Oswaldo Cruz 2018, 113, e180050. [Google Scholar] [CrossRef]
- Masur, H.; Brooks, J.T.; Benson, C.A.; Holmes, K.K.; Pau, A.K.; Kaplan, J.E. Prevention and Treatment of Opportunistic Infections in HIV-Infected Adults and Adolescents: Updated Guidelines from the Centers for Disease Control and Prevention, National Institutes of Health, and HIV Medicine Association of the Infectious Diseases Society of America. Clin. Infect. Dis. 2014, 58, 1308–1311. [Google Scholar]
- Setianingrum, F.; Rautemaa-Richardson, R.; Denning, D.W. Pulmonary Cryptococcosis: A Review of Pathobiology and Clinical Aspects. Med. Mycol. 2019, 57, 133–150. [Google Scholar] [CrossRef]
- Rajasingham, R.; Rolfes, M.A.; Birkenkamp, K.E.; Meya, D.B.; Boulware, D.R. Cryptococcal Meningitis Treatment Strategies in Resource-Limited Settings: A Cost-Effectiveness Analysis. PLoS Med. 2012, 9, e1001316. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.; Slechta, E.S.; Gates-Hollingsworth, M.A.; Neary, B.; Barker, A.P.; Bauman, S.; Kozel, T.R.; Hanson, K.E. Large-Scale Evaluation of the Immuno-Mycologics Lateral Flow and Enzyme-Linked Immunoassays for Detection of Cryptococcal Antigen in Serum and Cerebrospinal Fluid. Clin. Vaccine Immunol. 2013, 20, 52–55. [Google Scholar] [CrossRef] [Green Version]
- Del Negro, G.M.B.; Hong, M.A.; Duarte, A.J.S.; Shikanai-Yasuda, M.A.; Batista, L.; Benard, G. Antigen-Specific Immunosuppression in Paracoccidioidomycosis. Am. J. Trop. Med. Hyg. 1996, 54, 7–12. [Google Scholar] [CrossRef]
- Kamikawa, C.M.; Mendes, R.P.; Vicentini, A.P. Standardization and Validation of Dot-ELISA Assay for Paracoccidioides brasiliensis Antibody Detection. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, A.L.E.; Oliveira, M.A.P.; Silva, L.O.S.; Inácio, M.M.; Bailão, A.M.; Parente-Rocha, J.A.; Cruz-Leite, V.R.M.; Paccez, J.D.; de Almeida Soares, C.M.; Weber, S.S.; et al. Immunoproteomic Approach of Extracellular Antigens from Paracoccidioides Species Reveals Exclusive B-Cell Epitopes. Front. Microbiol. 2020, 10, 2968. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.D.M.; Patané, J.S.L.; Taylor, M.L.; Gómez, B.L.; Theodoro, R.C.; de Hoog, S.; Engelthaler, D.M.; Zancopé-Oliveira, R.M.; Felipe, M.S.S.; Barker, B.M. Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma. PLoS Negl. Trop. Dis. 2016, 10, e0004732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oladele, R.O.; Ayanlowo, O.O.; Richardson, M.D.; Denning, D.W. Histoplasmosis in Africa: An Emerging or a Neglected Disease? PLoS Negl. Trop. Dis. 2018, 12, e0006046. [Google Scholar] [CrossRef]
- Bahr, N.C.; Antinori, S.; Wheat, J.; Sarosi, G.A. Histoplasmosis Infections Worldwide: Thinking Outside of the Ohio River Valley. Curr. Trop. Med. Rep. 2016, 176, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Garfoot, A.L.; Reppleye, C.A. Histoplasma capsulatum Surmounts Obstacles to Intracellular Pathogenesis. FEBS J. 2016, 283, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Ray, S.C.; Rappleye, C.A. Flying under the Radar: Histoplasma capsulatum Avoidance of Innate Immune Recognition. Semin. Cell Dev. Biol. 2018, 89, 91–98. [Google Scholar] [CrossRef]
- Kauffman, C.A. Histoplasmosis: A Clinical and Laboratory Update. Clin. Microbiol. Rev. 2007, 20, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Wheat, L.J.; Azar, M.M.; Bahr, N.C.; Spec, A.; Relich, R.F.; Hage, C. Histoplasmosis. Infect. Dis. Clin. N. Am. 2016, 30, 207–227. [Google Scholar] [CrossRef]
- Connolly, P.A.; Durkin, M.M.; Lemonte, A.M.; Hackett, E.J.; Wheat, L.J. Detection of Histoplasma Antigen by a Quantitative Enzyme Immunoassay. Clin. Vaccine Immunol. 2007, 14, 1587–1591. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, A.J.; Nosanchuk, J.D.; Zancopé-Oliveira, R.M. Diagnosis of Histoplasmosis. Braz. J. Microbiol. 2006, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.M.; Gómez, B.L. Diagnostic Methods for Histoplasmosis: Focus on Endemic Countries with Variable Infrastructure Levels. Curr. Trop. Med. Rep. 2014, 1, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hage, C.A.; Davis, T.E.; Fuller, D.; Egan, L.; Witt, J.R.; Wheat, L.J.; Knox, K.S. Diagnosis of Histoplasmosis by Antigen Detection in BAL Fluid. Chest 2010, 137, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.L.; Tobn, A.; Restrepo, A.; Queiroz-Telles, F.; Nucci, M. Epidemiology of Endemic Systemic Fungal Infections in Latin America. Med. Mycol. 2011, 49, 785–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.; Benedict, K.; Park, B.J.; Thompson, G.R. Coccidioidomycosis: Epidemiology. Clin. Epidemiol. 2013, 5, 185–197. [Google Scholar] [PubMed] [Green Version]
- Sunenshine, R.H.; Anderson, S.; Erhart, L.; Vossbrink, A.; Kelly, P.C.; Engelthaler, D.; Komatsu, K. Public Health Surveillance for Coccidioidomycosis in Arizona. Ann. N. Y. Acad. Sci. 2007, 1111, 96–102. [Google Scholar] [CrossRef]
- Laniado-Laborín, R.; Arathoon, E.G.; Canteros, C.; Muñiz-Salazar, R.; Rendon, A. Coccidioidomycosis in Latin America. Med. Mycol. 2019, 57, S46–S55. [Google Scholar] [CrossRef]
- Giacomazzi, J.; Baethgen, L.; Carneiro, L.C.; Millington, M.A.; Denning, D.W.; Colombo, A.L.; Pasqualotto, A.C. The Burden of Serious Human Fungal Infections in Brazil. Mycoses 2016, 59, 145–150. [Google Scholar] [CrossRef]
- de Aguiar Cordeiro, R.; Brilhante, R.S.N.; Rocha, M.F.G.; Bandeira, S.P.; Fechine, M.A.B.; de Camargo, Z.P.; Sidrim, J.J.C. Twelve Years of Coccidioidomycosis in Ceará State, Northeast Brazil: Epidemiologic and Diagnostic Aspects. Diagn. Microbiol. Infect. Dis. 2010, 66, 65–72. [Google Scholar] [CrossRef]
- Kirkland, T.N.; Fierer, J. Coccidioides immitis and posadasii; a Review of Their Biology, Genomics, Pathogenesis, and Host Immunity. Virulence 2018, 9, 1426–1435. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C.; Barker, B.M.; Hoover, S.; Nix, D.E.; Ampel, N.M.; Frelinger, J.A.; Orbach, M.J.; Galgiani, J.N. Recent Advances in Our Understanding of the Environmental, Epidemiological, Immunological, and Clinical Dimensions of Coccidioidomycosis. Clin. Microbiol. Rev. 2013, 26, 505–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabe, L.M.; Malo, J.; Knox, K.S. Diagnosis and Management of Coccidioidomycosis. Clin. Chest Med. 2017, 38, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Malo, J.; Luraschi-Monjagatta, C.; Wolk, D.M.; Thompson, R.; Hage, C.A.; Knox, K.S. Update on the Diagnosis of Pulmonary Coccidioidomycosis. Ann. Am. Thorac. Soc. 2014, 11, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Malo, J.; Holbrook, E.; Zangeneh, T.; Strawter, C.; Oren, E.; Robey, I.; Erickson, H.; Carranza-Chahal, R.; Durkin, M.; Thompson, C.; et al. Comparison of Three Anti-Coccidioides Antibody Enzyme Immunoassay Kits for the Diagnosis of Coccidioidomycosis. Med. Mycol. 2020, 58, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.E.; Coakley, B.; Santelli, A.C.; Hentz, J.G.; Wengenack, N.L. Serologic Testing for Symptomatic Coccidioidomycosis in Immunocompetent and Immunosuppressed Hosts. Mycopathologia 2006, 162, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Pappagianis, D. Serologic Studies in Coccidioidomycosis. Semin. Respir. Infect. 2001, 16, 242–250. [Google Scholar] [CrossRef]
- Grill, F.J.; Grys, T.E.; Grill, M.F.; Roeder, A.; Blair, J.E.; Lake, D.F. Development of a Quantitative Antigen Assay to Detect Coccidioidal Chitinase-1 (CTS1) in Human Serum. Open Forum Infect. Dis. 2021, 8, ofab344. [Google Scholar] [CrossRef]
- Latgé, J.-P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2020, 33, e00140-18. [Google Scholar] [CrossRef]
- Mccormick, A.; Loeffler, J.; Ebel, F. Aspergillus fumigatus: Contours of an Opportunistic Human Pathogen. Cell. Microbiol. 2010, 12, 1535–1543. [Google Scholar] [CrossRef]
- Krishnan, S.; Manavathu, E.K.; Chandrasekar, P.H. Aspergillus flavus: An Emerging Non-fumigatus Aspergillus Species of Significance. Mycoses 2009, 52, 206–222. [Google Scholar] [CrossRef]
- Henriet, S.S.V.; Verweij, P.E.; Warris, A. Aspergillus nidulans and Chronic Granulomatous Disease: A Unique Host-Pathogen Interaction. J. Infect. Dis. 2012, 206, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Person, A.K.; Chudgar, S.M.; Norton, B.L.; Tong, B.C.; Stout, J.E. Aspergillus niger: An Unusual Cause of Invasive Pulmonary Aspergillosis. J. Med. Microbiol. 2010, 59, 834–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastor, F.J.; Guarro, J. Treatment of Aspergillus terreus Infections: A Clinical Problem Not yet Resolved. Int. J. Antimicrob. Agents 2014, 44, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Kousha, M.; Tadi, R.; Soubani, A.O. Pulmonary Aspergillosis: A Clinical Review. Eur. Respir. Rev. 2011, 20, 156–174. [Google Scholar] [CrossRef] [Green Version]
- Kontoyiannis, D.; Bodey, G. Invasive Aspergillosis in 2002: An Update. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 161–172. [Google Scholar] [CrossRef]
- Kanj, A.; Abdallah, N.; Soubani, A.O. The Spectrum of Pulmonary Aspergillosis. Respir. Med. 2018, 141, 121–131. [Google Scholar] [CrossRef]
- Godet, C.; Alastruey-Izquierdo, A.; Flick, H.; Hennequin, C.; Mikilps-Mikgelbs, R.; Munteanu, O.; Page, I.; Seidel, D.; Salzer, H.J.F. A CPAnet Consensus Statement on Research Priorities for Chronic Pulmonary Aspergillosis: A Neglected Fungal Infection That Requires Attention. J. Antimicrob. Chemother. 2018, 73, 280–286. [Google Scholar] [CrossRef]
- Niu, Y.; Li, J.; Shui, W.; Li, D.; Yu, C.; Fu, X.; Zhang, C. Clinical Features and Outcome of Patients with Chronic Pulmonary Aspergillosis in China: A Retrospective, Observational Study. J. De Mycol. Médicale 2020, 30, 101041. [Google Scholar] [CrossRef]
- Shah, A.; Armstrong-James, D.; Chotirmall, S.H. Respiratory Mycoses: A Call to Action to Recognize, Educate and Invest. Mycopathologia 2021, 186, 569–573. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nosanchuk, J.D. Fungal Diseases as Neglected Pathogens: A Wake-up Call to Public Health Officials. PLOS Negl. Trop. Dis. 2020, 14, e0007964. [Google Scholar] [CrossRef] [Green Version]
- Bongomin, F.; Gago, S.; Oladele, R.; Denning, D. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, T.F.; Thompson, G.R.; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef] [PubMed]
- Tortorano, A.M.; Esposto, M.C.; Prigitano, A.; Grancini, A.; Ossi, C.; Cavanna, C.; Cascio, G. lo Cross-Reactivity of Fusarium spp. in the Aspergillus Galactomannan Enzyme-Linked Immunosorbent Assay. J. Clin. Microbiol. 2012, 50, 1051–1053. [Google Scholar] [CrossRef] [Green Version]
- Wheat, L.J.; Hackett, E.; Durkin, M.; Connolly, P.; Petraitiene, R.; Walsh, T.J.; Knox, K.; Hage, C. Histoplasmosis-Associated Cross-Reactivity in the BioRad Platelia Aspergillus Enzyme Immunoassay. Clin. Vaccine Immunol. 2007, 14, 638–640. [Google Scholar] [CrossRef] [Green Version]
- Fraczek, M.G.; Kirwan, M.B.; Moore, C.B.; Morris, J.; Denning, D.W.; Richardson, M.D. Volume Dependency for Culture of Fungi from Respiratory Secretions and Increased Sensitivity of Aspergillus Quantitative PCR. Mycoses 2014, 57, 69–78. [Google Scholar] [CrossRef]
- Richardson, M.; Page, I. Role of Serological Tests in the Diagnosis of Mold Infections. Curr. Fungal Infect. Rep. 2018, 12, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Pasqualotto, A.C. Aspergillosis: From Diagnosis to Prevention; Springer: Dordrecht, The Netherlands, 2010; ISBN 9789048124084. [Google Scholar]
- Chapman, N.; Abela-Oversteegen, L.; Doubell, A.; Chowdhary, V.; Gurjav, U.; Ong, M. Neglected Disease Research and Development: A Pivotal Moment for Global Health. 2017. Available online: http://www.finddx.org/wp-content/uploads/2017/02/GF_report16_all_web.pdf (accessed on 20 February 2022).
- Molloy, S.F.; Chiller, T.; Greene, G.S.; Burry, J.; Govender, N.P.; Kanyama, C.; Mfinanga, S.; Lesikari, S.; Mapoure, Y.N.; Kouanfack, C.; et al. Cryptococcal Meningitis: A Neglected NTD? PLoS Negl. Trop. Dis. 2017, 11, e0005575. [Google Scholar] [CrossRef] [Green Version]
- Kwon-Chung, K.J.; Bennett, J.E.; Wickes, B.L.; Meyer, W.; Cuomo, C.A.; Wollenburg, K.R.; Bicanic, T.A.; Castañeda, E.; Chang, Y.C.; Chen, J.; et al. The Case for Adopting the “Species Complex” Nomenclature for the Etiologic Agents of Cryptococcosis. mSphere 2017, 2, e00357-16. [Google Scholar] [CrossRef] [Green Version]
- Maziarz, E.K.; Perfect, J.R. Cryptococcosis. Infect. Dis. Clin. N. Am. 2016, 30, 179–206. [Google Scholar] [CrossRef] [Green Version]
- Cogliati, M.; Desnos-Ollivier, M.; McCormick-Smith, I.; Rickerts, V.; Ferreira-Paim, K.; Meyer, W.; Boekhout, T.; Hagen, F.; Theelen, B.; Inácio, J.; et al. Genotypes and Population Genetics of Cryptococcus neoformans and Cryptococcus gattii Species Complexes in Europe and the Mediterranean Area. Fungal Genet. Biol. 2019, 129, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lin, P.C.; Ye, J.R.; Su, S.S.; Dong, L.; Wu, Q.; Xu, H.Y.; Xie, Y.P.; Li, Y.P. The Performance of Serum Cryptococcal Capsular Polysaccharide Antigen Test, Histopathology and Culture of the Lung Tissue for Diagnosis of Pulmonary Cryptococcosis in Patients without HIV Infection. Infect. Drug Resist. 2018, 11, 2483–2490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazzoni, A.F.; Severo, C.B.; Barra, M.B.; Severo, L.C. Atypical Micromorphology and Uncommon Location of Cryptococcosis: A Histopathologic Study Using Special Histochemical Techniques (One Case Report). Mycopathologia 2009, 167, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Gazzoni, A.F.; Pegas, K.L.; Severo, L.C. Histopathological Techniques for Diagnosing Cryptococcosis Due to Capsule-Deficient Cryptococcus: Case Report. Rev. Soc. Bras. Med. Trop. 2008, 41, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Firacative, C.; Trilles, L.; Meyer, W. MALDI-TOF MS Enables the Rapid Identification of the Major Molecular Types within the Cryptococcus neoformans/C. gattii Species Complex. PLoS ONE 2012, 7, e37566. [Google Scholar] [CrossRef] [Green Version]
- McTaggart, L.R.; Lei, E.; Richardson, S.E.; Hoang, L.; Fothergill, A.; Zhang, S.X. Rapid Identification of Cryptococcus neoformans and Cryptococcus gattii by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2011, 49, 3050–3053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, N.; Gray, K.; Watera, C.; Nakiyingi, J.; Lugada, E.; Moore, M.; Lalloo, D.; Whitworth, J.A.G.; Gilks, C.F. Cryptococcal Infection in a Cohort of HIV-1-Infected Ugandan Adults. Aids 2002, 16, 1031–1038. [Google Scholar] [CrossRef]
- Kwizera, R.; Omali, D.; Tadeo, K.; Kasibante, J.; Rutakingirwa, M.K.; Kagimu, E.; Ssebambulidde, K.; Williams, D.A.; Rhein, J.; Boulware, D.; et al. Evaluation of the Dynamiker Cryptococcal Antigen Lateral Flow Assay for the Diagnosis of HIV-Associated Cryptococcosis. J. Clin. Microbiol. 2021, 59, e02421-20. [Google Scholar] [CrossRef]
- Likasitwattanakul, S.; Poneprasert, B.; Sirisanthana, V. Cryptococcosis in HIV-Infected Children. Southeast Asian J. Trop. Med. Public Health 2004, 35, 935–939. [Google Scholar]
- Vidal, J.E.; Boulware, D.R. Lateral Flow Assay for Cryptococcal Antigen: An Important Advance to Improve the Continuum of HIV Care and Reduce Cryptococcal Meningitis-Related Mortality. Rev. Inst. Med. Trop. Sao Paulo 2015, 57, 38–45. [Google Scholar] [CrossRef]
- Rolfes, M.; Butler, E.; von Hohenberg, M.; Nabeta, H.; Kwizera, R.; Rajasingham, R.; Bahr, N.; Meya, D.; Boulware, D. Evaluation of a Novel Point-of-Care Lateral Flow Assay to Detect Cryptococcal Antigen in Plasma and CSF. Poster #953, Session 159. In Proceedings of the CROI, Seattle, WA, USA, 8 March 2012; p. 7. [Google Scholar]
- Vijayan, T.; Bauman, S.; Chiller, T.; Klausner, J. Test Performance of a Novel Lateral-Flow Assay to Detect Cryptococcal Disease. 2012. Available online: https://idsa.confex.com/idsa/2012/webprogram/Paper37789.html (accessed on 20 February 2022).
- Huang, H.R.; Fan, L.C.; Rajbanshi, B.; Xu, J.F. Evaluation of a New Cryptococcal Antigen Lateral Flow Immunoassay in Serum, Cerebrospinal Fluid and Urine for the Diagnosis of Cryptococcosis: A Meta-Analysis and Systematic Review. PLoS ONE 2015, 10, e0127117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, J.E.; Toniolo, C.; Paulino, A.; Colombo, A.L.; Martins, M.D.A.; da Silva Meira, C.; Azevedo, R.G.S.; Pereira-Chioccola, V.L.; Gomes, H.R.; Dos Santos Lazera, M.; et al. Performance of Cryptococcal Antigen Lateral Flow Assay in Serum, Cerebrospinal Fluid, Whole Blood, and Urine in HIV-Infected Patients with Culture-Proven Cryptococcal Meningitis Admitted at a Brazilian Referral Center. Rev. Inst. Med. Trop. Sao Paulo 2018, 60. [Google Scholar] [CrossRef]
- Magambo, K.A.; Kalluvya, S.E.; Kapoor, S.W.; Seni, J.; Chofle, A.A.; Fitzgerald, D.W.; Downs, J.A. Utility of Urine and Serum Lateral Flow Assays to Determine the Prevalence and Predictors of Cryptococcal Antigenemia in HIV-Positive Outpatients Beginning Antiretroviral Therapy in Mwanza, Tanzania. J. Int. AIDS Soc. 2014, 17, 19040. [Google Scholar] [CrossRef] [PubMed]
- Longley, N.; Jarvis, J.N.; Meintjes, G.; Boulle, A.; Cross, A.; Kelly, N.; Govender, N.P.; Bekker, L.G.; Wood, R.; Harrison, T.S. Cryptococcal Antigen Screening in Patients Initiating ART in South Africa: A Prospective Cohort Study. Clin. Infect. Dis. 2016, 62, 581–587. [Google Scholar] [CrossRef] [Green Version]
- Mfinanga, S.; Chanda, D.; Kivuyo, S.L.; Guinness, L.; Bottomley, C.; Simms, V.; Chijoka, C.; Masasi, A.; Kimaro, G.; Ngowi, B.; et al. Cryptococcal Meningitis Screening and Community-Based Early Adherence Support in People with Advanced HIV Infection Starting Antiretroviral Therapy in Tanzania and Zambia: An Open-Label, Randomised Controlled Trial. Lancet 2015, 385, 2173–2182. [Google Scholar] [CrossRef]
- Letang, E.; Müller, M.C.; Ntamatungiro, A.J.; Kimera, N.; Faini, D.; Furrer, H.; Battegay, M.; Tanner, M.; Hatz, C.; Boulware, D.R.; et al. Cryptococcal Antigenemia in Immunocompromised Human Immunodeficiency Virus Patients in Rural Tanzania: A Preventable Cause of Early Mortality. Open Forum Infect. Dis. 2015, 2, ofv046. [Google Scholar] [CrossRef]
- Pac, L.; Horwitz, M.M.; Namutebi, A.M.; Auerbach, B.J.; Semeere, A.; Namulema, T.; Schwarz, M.; Bbosa, R.; Muruta, A.; Meya, D.B.; et al. Implementation and Operational Research: Integrated Pre-Antiretroviral Therapy Screening and Treatment for Tuberculosis and Cryptococcal Antigenemia. J. Acquir. Immune Defic. Syndr. 2015, 68, e69–e76. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.E.; Vallabhaneni, S.; Smith, R.M.; Chideya-Chihota, S.; Chehab, J.; Park, B. Cryptococcal Antigen Screening and Early Antifungal Treatment to Prevent Cryptococcal Meningitis: A Review of the Literature. J. Acquir. Immune Defic. Syndr. 2015, 68, S331–S339. [Google Scholar] [CrossRef]
- Camargo, Z.P.; Taborda, C.P.; Rodrigues, E.G.; Travassos, L.R. The Use of Cell-Free Antigens of Paracoccidioides brasiliensis in Serological Tests. Med. Mycol. 1991, 29, 31–38. [Google Scholar] [CrossRef]
- Sá-Nunes, A.; Medeiros, A.I.; Nicolete, R.; Frantz, F.G.; Panunto-Castelo, A.; Silva, C.L.; Faccioli, L.H. Efficacy of Cell-Free Antigens in Evaluating Cell Immunity and Inducing Protection in a Murine Model of Histoplasmosis. Microbes Infect. 2005, 7, 584–592. [Google Scholar] [CrossRef]
- dos Santos, D.F.; Nicolete, R.; de Souza, P.R.M.; Bitencourt, C.D.S.; dos Santos Junior, R.R.; Bonato, V.L.D.; Silva, C.L.; Faccioli, L.H. Characterization and In Vitro Activities of Cell-Free Antigens from Histoplasma capsulatum-Loaded Biodegradable Microspheres. Eur. J. Pharm. Sci. 2009, 38, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Ferreira Fernandes, G.; Oliveira dos Santos, P.; Messias Rodrigues, A.; Augusto Sasaki, A.; Burger, E.; Pires de Camargo, Z. Characterization of Virulence Profile, Protein Secretion and Immunogenicity of Different Sporothrix schenckii sensu stricto Isolates Compared with S. globosa and S. brasiliensis Species. Virulence 2013, 4, 241–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holbrook, E.D.; Kemski, M.M.; Richer, S.M.; Wheat, L.J.; Rappleye, C.A. Glycosylation and Immunoreactivity of the Histoplasma capsulatum Cfp4 Yeast-Phase Exoantigen. Infect. Immun. 2014, 82, 4414–4425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, A.J.; Bartholomew, M.A.; Figueroa, J.; Fenelon, L.E.; Hay, R.J. Production of Species-Specific Murine Monoclonal Antibodies against Cryptococcus neoformans which Recognize a Noncapsular Exoantigen. J. Clin. Microbiol. 1991, 29, 980–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida-Paes, R.; Pimenta, M.A.; Pizzini, C.V.; Monteiro, P.C.F.; Peralta, J.M.; Nosanchuk, J.D.; Zancopé-Oliveira, R.M. Use of Mycelial-Phase Sporothrix schenckii Exoantigens in an Enzyme-Linked Immunosorbent Assay for Diagnosis of Sporotrichosis by Antibody Detection. Clin. Vaccine Immunol. 2007, 14, 244–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, S.S.; Parente, A.F.A.; Borges, C.L.; Parente, J.A.; Bailão, A.M.; de Almeida Soares, C.M. Analysis of the Secretomes of Paracoccidioides Mycelia and Yeast Cells. PLoS ONE 2012, 7, e52470. [Google Scholar] [CrossRef] [PubMed]
- Rigobello, F.F.; Marquez, A.S.; Lopes, J.D.; Nakanishi-Ito, F.A.; Itano, E.N. Patients with Chronic-Form Paracoccidioidomycosis Present High Serum Levels of IgE Anti-Paracoccidioides brasiliensis Gp70. Mycopathologia 2013, 175, 307–313. [Google Scholar] [CrossRef]
- Maia, D.C.G.; Gonçalves, A.C.; Ferreira, L.S.; Manente, F.A.; Portuondo, D.L.; Vellosa, J.C.R.; Polesi, M.C.; Batista-Duharte, A.; Carlos, I.Z. Response of Cytokines and Hydrogen Peroxide to Sporothrix schenckii Exoantigen in Systemic Experimental Infection. Mycopathologia 2016, 181, 207–215. [Google Scholar] [CrossRef] [Green Version]
- della Terra, P.P.; Rodrigues, A.M.; Fernandes, G.F.; Nishikaku, A.S.; Burger, E.; de Camargo, Z.P. Exploring Virulence and Immunogenicity in the Emerging Pathogen Sporothrix Brasiliensis. PLoS Negl. Trop. Dis. 2017, 11, e0005903. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.S.; Miranda, K.; Silva, L.S.; Freymüller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The Pathogenic Fungus Paracoccidioides brasiliensis Exports Extracellular Vesicles Containing Highly Immunogenic α-Galactosyl Epitopes. Eukaryot. Cell 2011, 10, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Lenhard-Vidal, A.; Assolini, J.P.; Ono, M.A.; Bredt, C.S.O.; Sano, A.; Itano, E.N. Paracoccidioides brasiliensis and P. lutzii Antigens Elicit Different Serum IgG Responses in Chronic Paracoccidioidomycosis. Mycopathologia 2013, 176, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, G.F.; Lopes-Bezerra, L.M.; Bernardes-Engemann, A.R.; Schubach, T.M.P.; Dias, M.A.G.; Pereira, S.A.; de Camargo, Z.P. Serodiagnosis of Sporotrichosis Infection in Cats by Enzyme-Linked Immunosorbent Assay Using a Specific Antigen, SsCBF, and Crude Exoantigens. Vet. Microbiol. 2011, 147, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.A.; Almeida-Paes, R.; Guimarães, A.J.; Valente, R.H.; Soares, C.M.D.A.; Zancopé-Oliveira, R.M. Immunoproteomics Reveals Pathogen’s Antigens Involved in Homo Sapiens—Histoplasma capsulatum Interaction and Specific Linear B-Cell Epitopes in Histoplasmosis. Front. Cell. Infect. Microbiol. 2020, 2020, 659. [Google Scholar] [CrossRef] [PubMed]
- Cavassani, K.A.; Tristao, F.S.M.; Oliveira, L.L.; Rocha, F.A.; Vancim, J.O.; Moreira, A.P.; Campanelli, A.P.; Panagio, L.A.; Milanezi, C.M.; Martinez, R.; et al. Cell-Free Antigens from Paracoccidioides brasiliensis Drive IL-4 Production and Increase the Severity of Paracoccidioidomycosis. PLoS ONE 2011, 6, e21423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Santos, D.F.; Bitencourt, C.S.; Gelfuso, G.M.; Pereira, P.A.T.; de Souza, P.R.M.; Sorgi, C.A.; Nicolete, R.; Faccioli, L.H. Biodegradable Microspheres Containing Leukotriene B4 and Cell-Free Antigens from Histoplasma capsulatum Activate Murine Bone Marrow-Derived Macrophages. Eur. J. Pharm. Sci. 2011, 44, 580–588. [Google Scholar] [CrossRef]
- Standard, P.G.; Kaufman, L. Specific Immunological Test for the Rapid Identification of Members of the Genus Histoplasma. J. Clin. Microbiol. 1976, 3, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Polonelli, L.; Morace, G. Exoantigen Studies of Sporothrix schenckii, Ceratocystis minor, and Graphium penicilliodes Cultures. J. Clin. Microbiol. 1982, 15, 362–365. [Google Scholar] [CrossRef] [Green Version]
- Ehrhard, H.B.; Pine, L. Factors Influencing the Production of H and M Antigens by Histoplasma capsulatum: Effect of Physical Factors and Composition of Medium. Appl. Microbiol. 1972, 23, 250–261. [Google Scholar] [CrossRef]
- Zancopé-Oliveira, R.M.; Bragg, S.L.; Reiss, E.; Peralta, J.M. Immunochemical Analysis of the H and M Glycoproteins from Histoplasma capsulatum. Clin. Diagn. Lab. Immunol. 1994, 1, 563–568. [Google Scholar] [CrossRef]
- Zancopé-Oliveira, R.M.; Reiss, E.; Lott, T.J.; Mayer, L.W.; Deepe, G.S. Molecular Cloning, Characterization, and Expression of the M Antigen of Histoplasma capsulatum. Infect. Immun. 1999, 67, 1947–1953. [Google Scholar] [CrossRef]
- Guimaräes, A.J.; Hamilton, A.J.; de M. Guedes, H.L.; Nosanchuk, J.D.; Zancopé-Oliveira, R.M. Biological Function and Molecular Mapping of M Antigen in Yeast Phase of Histoplasma capsulatum. PLoS ONE 2008, 3, e3449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, V.C.; Martins, E.M.N.; Boeloni, J.N.; Coitinho, J.B.; Serakides, R.; Goes, A.M. Additive Effect of RPb27 Immunization and Chemotherapy in Experimental Paracoccidioidomycosis. PLoS ONE 2011, 6, e17885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo, Z.P.; Berzaghi, R.; Amaral, C.C.; Silva, S.H.M. Simplified Method for Producing Paracoccidioides brasiliensis Exoantigens for Use in Immunodiffusion Tests. Med. Mycol. 2003, 41, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, A.P.V. Paracoccidioidomicose: Histórico, Etiologia, Epidemiologia, Patogênese, Formas Clínicas, Diagnóstico Laboratorial e Antígenos. BEPA Bol. Epidemiol. Paul. 2008, 5, 1–17. [Google Scholar]
- Jungblut, P.R. Proteome Analysis of Bacterial Pathogens. Microbes. Infect. 2001, 3, 831–840. [Google Scholar] [CrossRef]
- Sylvestre, T.F.; Cavalcante, R.D.S.; da Silva, J.D.F.; Paniago, A.M.M.; Weber, S.S.; Pauletti, B.A.; Carvalho, L.R.D.; dos Santos, L.D.; Mendes, R.P. Serological Proteomic Biomarkers to Identify Paracoccidioides Species and Risk of Relapse. PLoS ONE 2018, 13, e0202804. [Google Scholar] [CrossRef] [Green Version]
- Hess, J.L.; Blazer, L.; Romer, T.; Faber, L.; Buller, R.M.; Boyle, M.D.P. Immunoproteomics. J. Chromatogr. B 2005, 815, 65–75. [Google Scholar] [CrossRef]
- Carmona, S.J.; Nielsen, M.; Schafer-Nielsen, C.; Mucci, J.; Altcheh, J.; Balouz, V.; Tekiel, V.; Frasch, A.C.; Campetella, O.; Buscaglia, C.A.; et al. Towards High-Throughput Immunomics for Infectious Diseases: Use of next-Generation Peptide Microarrays for Rapid Discovery and Mapping of Antigenic Determinants. Mol. Cell. Proteom. 2015, 14, 1871–1884. [Google Scholar] [CrossRef] [Green Version]
- Gaseitsiwe, S.; Valentini, D.; Mahdavifar, S.; Reilly, M.; Ehrnst, A.; Maeurer, M. Peptide Microarray-Based Identification of Mycobacterium Tuberculosis Epitope Binding to HLA-DRB1*0101, DRB1*1501, and DRB1*0401. Clin. Vaccine Immunol. 2010, 17, 168–175. [Google Scholar] [CrossRef] [Green Version]
- Luu, L.D.W.; Octavia, S.; Aitken, C.; Zhong, L.; Raftery, M.J.; Sintchenko, V.; Lan, R. Surfaceome Analysis of Australian Epidemic Bordetella Pertussis Reveals Potential Vaccine Antigens. Vaccine 2020, 38, 539–548. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving Live Organisms for a Fast Proteomic Identification of Surface Proteins. J. Proteom. 2014, 97, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science (1979) 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Liu, I.-J.; Lu, R.-M.; Wu, H.-C. Advancement and Applications of Peptide Phage Display Technology in Biomedical Science. J. Biomed. Sci. 2016, 23, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Parray, H.A.; Shrivastava, T.; Sinha, S.; Luthra, K. Phage Display Antibody Libraries: A Robust Approach for Generation of Recombinant Human Monoclonal Antibodies. Int. J. Biol. Macromol. 2019, 135, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.A.; Eweida, A.E.; Sheweita, S.A. B-Cell Epitope Mapping for the Design of Vaccines and Effective Diagnostics. Trials Vaccinol. 2016, 5, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Flower, D.R. Immunoinformatics. Predicting Immunogenicity in Silico. Preface. Methods Mol. Biol. 2007, 409, v–vi. [Google Scholar]
- Queiroz, L.D.P.; de Camargo, Z.P.; Tadano, T.; Rodrigues, A.M.; Takarara, D.T.; Gegembauer, G.; Araujo, L.M.; Hahn, R.C. Serological and Antigenic Profiles of Clinical Isolates of Paracoccidioides spp. from Central Western Brazil. Mycoses 2014, 57, 466–472. [Google Scholar] [CrossRef]
- Gautam, P.; Sundaram, C.S.; Madan, T.; Gade, W.N.; Shah, A.; Sirdeshmukh, R.; Sarma, P.U. Identification of Novel Allergens of Aspergillus fumigatus Using Immunoproteomics Approach. Clin. Exp. Allergy 2007, 37, 1239–1249. [Google Scholar] [CrossRef]
- Tarcha, E.J.; Basrur, V.; Hung, C.Y.; Gardner, M.J.; Cole, G.T. Multivalent Recombinant Protein Vaccine against Coccidioidomycosis. Infect. Immun. 2006, 74, 5802–5813. [Google Scholar] [CrossRef] [Green Version]
- Virginio, E.D.; Kubitschek-Barreira, P.H.; Batista, M.V.; Schirmer, M.R.; Abdelhay, E.; Shikanai-Yasuda, M.A.; Lopes-Bezerra, L.M. Immunoproteome of Aspergillus fumigatus Using Sera of Patients with Invasive Aspergillosis. Int. J. Mol. Sci. 2014, 15, 14505–14530. [Google Scholar] [CrossRef] [Green Version]
- Bär, E.; Gladiator, A.; Bastidas, S.; Roschitzki, B.; Acha-Orbea, H.; Oxenius, A.; LeibundGut-Landmann, S. A Novel Th Cell Epitope of Candida Albicans Mediates Protection from Fungal Infection. J. Immunol. 2012, 188, 5636–5643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulton, K.M.; Baltat, I.; Twine, S.M. Immunoproteomics Methods and Techniques. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 2024, pp. 25–58. [Google Scholar]
- Martins, L.M.S.; Andrade, H.M.D.; Vainstein, M.H.; Wanke, B.; Schrank, A.; Balaguez, C.B.; Santos, P.R.D.; Santi, L.; Pires, S.D.F.; da Silva, A.S.; et al. Immunoproteomics and Immunoinformatics Analysis of Cryptococcus gattii: Novel Candidate Antigens for Diagnosis. Future Microbiol. 2013, 8, 549–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, A.M.; Kubitschek-Barreira, P.H.; Fernandes, G.F.; de Almeida, S.R.; Lopes-Bezerra, L.M.; de Camargo, Z.P. Immunoproteomic Analysis Reveals a Convergent Humoral Response Signature in the Sporothrix schenckii Complex. J. Proteom. 2015, 115, 8–22. [Google Scholar] [CrossRef]
- Herbert, B.R.; Grinyer, J.; McCarthy, J.T.; Isaacs, M.; Harry, E.J.; Nevalainen, H.; Traini, M.D.; Hunt, S.; Schulz, B.; Laver, M.; et al. Improved 2-DE of Microorganisms after Acidic Extraction. Electrophoresis 2006, 27, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Souza, G.H.M.F.; Garcia, J.S.; Rech, E.L. Detection and Expression Analysis of Recombinant Proteins in Plant-Derived Complex Mixtures Using NanoUPLC-MSE. J. Sep. Sci. 2011, 34, 2618–2630. [Google Scholar] [CrossRef] [Green Version]
- Murad, A.M.; Rech, E.L. NanoUPLC-MSE Proteomic Data Assessment of Soybean Seeds Using the Uniprot Database. BMC Biotechnol. 2012, 12, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitarch, A.; Nombela, C.; Gil, C. Candida Albicans Biology and Pathogenicity: Insights from Proteomics. In Microbial Proteomics: Functional Biology of Whole Organisms; Wiley Blackwell: Hoboken, NJ, USA, 2005; pp. 285–330. ISBN 9780471973164. [Google Scholar]
- Lee, P.Y.; Gam, L.H.; Yong, V.C.; Rosli, R.; Ng, K.P.; Chong, P.P. Immunoproteomic Analysis of Antibody Response to Cell Wall-Associated Proteins of Candida Tropicalis. J. Appl. Microbiol. 2014, 117, 854–865. [Google Scholar] [CrossRef]
- Jobbins, S.E.; Hill, C.J.; D’Souza-Basseal, J.M.; Padula, M.P.; Herbert, B.R.; Krockenberger, M.B. Immunoproteomic Approach to Elucidating the Pathogenesis of Cryptococcosis Caused by Cryptococcus gattii. J. Proteome Res. 2010, 9, 3832–3841. [Google Scholar] [CrossRef]
- Mini, R.; Bernardini, G.; Salzano, A.M.; Renzone, G.; Scaloni, A.; Figura, N.; Santucci, A. Comparative Proteomics and Immunoproteomics of Helicobacter Pylori Related to Different Gastric Pathologies. J. Chromatogr. B 2006, 833, 63–79. [Google Scholar] [CrossRef]
- Pedersen, S.K.; Sloane, A.J.; Prasad, S.S.; Sebastian, L.T.; Lindner, R.A.; Hsu, M.; Robinson, M.; Bye, P.T.; Weinberger, R.P.; Harry, J.L. An Immunoproteomic Approach for Identification of Clinical Biomarkers for Monitoring Disease: Application to Cystic Fibrosis. Mol. Cell. Proteom. 2005, 4, 1052–1060. [Google Scholar] [CrossRef] [Green Version]
- Shin, G.W.; Palaksha, K.J.; Kim, Y.R.; Nho, S.W.; Cho, J.H.; Heo, N.E.; Heo, G.J.; Park, S.C.; Jung, T.S. Immunoproteomic Analysis of Capsulate and Non-Capsulate Strains of Lactococcus Garvieae. Vet. Microbiol. 2007, 119, 205–212. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Cordeiro, R.; Patoilo, K.R.N.; Praciano, S.B.; Medrano, D.J.A.; de Farias Marques, F.J.; Martins, L.M.S.; Eulalio, K.D.; de Deus Filho, A.; do Amparo Salmito Cavalvanti, M.; Fechine, M.A.B.; et al. Antigens of Coccidioides posadasii as an Important Tool for the Immunodiagnosis of Coccidioidomycosis. Mycopathologia 2013, 175, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarcha, E.J.; Basrur, V.; Hung, C.Y.; Gardner, M.J.; Cole, G.T. A Recombinant Aspartyl Protease of Coccidioides posadasii Induces Protection against Pulmonary Coccidioidomycosis in Mice. Infect. Immun. 2006, 74, 516–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.-J.; Holbrook, E.; Liao, Y.-R.; Zarnowski, R.; Andes, D.R.; Wheat, L.J.; Malo, J.; Hung, C.-Y. Characterization of an Uncinocarpus Reesii-Expressed Recombinant Tube Precipitin Antigen of Coccidioides posadasii for Serodiagnosis. PLoS ONE 2019, 14, e0221228. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Kubitschek-Barreira, P.H.; Pinheiro, B.G.; Teixeira-Ferreira, A.; Hahn, R.C.; de Camargo, Z.P. Immunoproteomic Analysis Reveals Novel Candidate Antigens for the Diagnosis of Paracoccidioidomycosis Due to Paracoccidioides lutzii. J. Fungi 2020, 6, 357. [Google Scholar] [CrossRef]
- Frank, R. Spot-Synthesis: An Easy Technique for the Positionally Addressable, Parallel Chemical Synthesis on a Membrane Support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Andresen, H.; Bier, F.F. Peptide Microarrays for Serum Antibody Diagnostics. Methods Mol. Biol. 2009, 509, 123–134. [Google Scholar]
- Jian, M.; Su, M.; Gao, J.; Wang, Z. Peptide Microarray-Based Fluorescence Assay for Quantitatively Monitoring the Tumor-Associated Matrix Metalloproteinase-2 Activity. Sens. Actuators B Chem. 2020, 304, 127320. [Google Scholar] [CrossRef]
- Svarovsky, S.A.; Gonzalez-Moa, M.J. High-Throughput Platform for Rapid Deployment of Antimicrobial Agents. ACS Comb. Sci. 2011, 13, 634–638. [Google Scholar] [CrossRef]
- Shah, P.; Wu, W.S.; Chen, C.S. Systematical Analysis of the Protein Targets of Lactoferricin B and Histatin-5 Using Yeast Proteome Microarrays. Int. J. Mol. Sci. 2019, 20, 4218. [Google Scholar] [CrossRef] [Green Version]
- Hernáez, M.L.; Ximénez-Embún, P.; Martínez-Gomariz, M.; Gutiérrez-Blázquez, M.D.; Nombela, C.; Gil, C. Identification of Candida Albicans Exposed Surface Proteins in Vivo by a Rapid Proteomic Approach. J. Proteom. 2010, 73, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Voltersen, V.; Blango, M.G.; Herrmann, S.; Schmidt, F.; Heinekamp, T.; Strassburger, M.; Krüger, T.; Bacher, P.; Lother, J.; Weiss, E.; et al. Proteome Analysis Reveals the Conidial Surface Protein CcpA Essential for Virulence of the Pathogenic Fungus Aspergillus fumigatus. mBio 2018, 9, 1557–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, H.C.; Michaloski, J.S.; da Silva, J.F.; Scorzoni, L.; de Paula e Silva, A.C.A.; Marcos, C.M.; Assato, P.A.; Yamazaki, D.S.; Fusco-Almeida, A.M.; Giordano, R.J.; et al. Peptides Derived from a Phage Display Library Inhibit Adhesion and Protect the Host against Infection by Paracoccidioides brasiliensis and Paracoccidioides lutzii. Front. Pharmacol. 2016, 7, 509. [Google Scholar] [CrossRef] [PubMed]
- Portes, L.D.S.; Kioshima, E.S.; de Camargo, Z.P.; Batista, W.L.; Xander, P. Subtractive Phage Display Selection for Screening and Identification of Peptide Sequences with Potential Use in Serodiagnosis of Paracoccidioidomycosis Caused by Paracoccidioides brasiliensis. Lett. Appl. Microbiol. 2017, 65, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wu, S.; Lin, J.; Cheng, L.; Zhou, J.; Xie, J.; Huang, K.; Wang, X.; Yu, Y.; Chen, Z.; et al. Nanoparticles Targeted against Cryptococcal Pneumonia by Interactions between Chitosan and Its Peptide Ligand. Nano Lett. 2018, 18, 6207–6213. [Google Scholar] [CrossRef]
- Noya, O.; Patarroyo, M.E.; Guzmán, F.; Alarcón De Noya, B. Immunodiagnosis of Parasitic Diseases with Synthetic Peptides. Curr. Protein Pept. Sci. 2003, 4, 299–308. [Google Scholar] [CrossRef]
- He, Y.; Rappuoli, R.; de Groot, A.S.; Chen, R.T. Emerging Vaccine Informatics. J. Biomed. Biotechnol. 2010, 2010, 218590. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Chen, H.; Zhuang, S.; Feng, X.; Fang, Y.; Tang, H.; Dai, R.; Tang, L.; Liu, J.; Ma, T.; et al. Immunodominant Regions Prediction of Nucleocapsid Protein for SARS-CoV-2 Early Diagnosis: A Bioinformatics and Immunoinformatics Study. Glob. Health 2020, 114, 463–470. [Google Scholar] [CrossRef]
- Lorenzo, M.A.; Pachón, D.; Maier, A.; Bermúdez, H.; Losada, S.; Toledo, M.; Pujol, F.H.; Alarcón de Noya, B.; Noya, O.; Serrano, M.L. Immunoinformatics and Pepscan Strategies on the Path of a Peptide-Based Serological Diagnosis of COVID-19. J. Immunol. Methods 2021, 495, 113071. [Google Scholar] [CrossRef]
- Phan, I.Q.; Subramanian, S.; Kim, D.; Murphy, M.; Pettie, D.; Carter, L.; Anishchenko, I.; Barrett, L.K.; Craig, J.; Tillery, L.; et al. In Silico Detection of SARS-CoV-2 Specific B-Cell Epitopes and Validation in ELISA for Serological Diagnosis of COVID-19. Sci. Rep. 2021, 11, 4290. [Google Scholar] [CrossRef]
- Carvalho, G.B.F.; Resende, D.M.; Siqueira, L.M.V.; Lopes, M.D.; Lopes, D.O.; Coelho, P.M.Z.; Teixeira-Carvalho, A.; Ruiz, J.C.; Fonseca, C.T. Selecting Targets for the Diagnosis of Schistosoma Mansoni Infection: An Integrative Approach Using Multi-Omic and Immunoinformatics Data. PLoS ONE 2017, 12, e0182299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javadi Mamaghani, A.; Fathollahi, A.; Spotin, A.; mehdi Ranjbar, M.; Barati, M.; Aghamolaie, S.; Karimi, M.; Taghipour, N.; Ashrafi, M.; Tabaei, S.J.S. Candidate Antigenic Epitopes for Vaccination and Diagnosis Strategies of Toxoplasma Gondii Infection: A Review. Microb. Pathog. 2019, 137, 103788. [Google Scholar] [CrossRef] [PubMed]
- Aghamolaei, S.; Kazemi, B.; Bandehpour, M.; Ranjbar, M.M.; Rouhani, S.; Javadi Mamaghani, A.; Tabaei, S.J.S. Design and Expression of Polytopic Construct of Cathepsin-L1, SAP-2 and FhTP16.5 Proteins of Fasciola hepatica. J. Helminthol. 2020, 94, e134. [Google Scholar] [CrossRef] [PubMed]
- Miles, S.; Navatta, M.; Dematteis, S.; Mourglia-Ettlin, G. Identification of Universal Diagnostic Peptide Candidates for Neglected Tropical Diseases Caused by Cestodes through the Integration of Multi-Genome-Wide Analyses and Immunoinformatic Predictions. Infect. Genet. Evol. 2017, 54, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Bai, Q.; Wu, X.; Li, H.; Shao, J.; Sun, M.; Jiang, H.; Zhang, J. Paper-Based ELISA Diagnosis Technology for Human Brucellosis Based on a Multiepitope Fusion Protein. PLoS Negl. Trop. Dis. 2021, 15, e0009695. [Google Scholar] [CrossRef]
- Faria, A.R.; Costa, M.M.; Giusta, M.S.; Grimaldi, G.; Penido, M.L.O.; Gazzinelli, R.T.; Andrade, H.M. High-Throughput Analysis of Synthetic Peptides for the Immunodiagnosis of Canine Visceral Leishmaniasis. PLoS Negl. Trop. Dis. 2011, 5, e1310. [Google Scholar] [CrossRef] [Green Version]
- Shirai, H.; Prades, C.; Vita, R.; Marcatili, P.; Popovic, B.; Xu, J.; Overington, J.P.; Hirayama, K.; Soga, S.; Tsunoyama, K.; et al. Antibody Informatics for Drug Discovery. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 2002–2015. [Google Scholar] [CrossRef]
- Galanis, K.A.; Nastou, K.C.; Papandreou, N.C.; Petichakis, G.N.; Iconomidou, V.A. Linear B-Cell Epitope Prediction: A Performance Review of Currently Available Methods. bioRxiv 2019, 833418. [Google Scholar] [CrossRef] [Green Version]
- Ansari, H.R.; Raghava, G.P. Identification of Conformational B-Cell Epitopes in an Antigen from Its Primary Sequence. Immunome Res. 2010, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhao, X.; Sun, P.; Gao, B.; Ma, Z. Conformational B-Cell Epitopes Prediction from Sequences Using Cost-Sensitive Ensemble Classifiers and Spatial Clustering. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Raghava, G.P.S. BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physico-Chemical Properties. Lect. Notes Comput. Sci. 2004, 3239, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved Method for Predicting Linear B-Cell Epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Raghava, G.P.S. Prediction of Continuous B-Cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Sweredoski, M.J.; Baldi, P. COBEpro: A Novel System for Predicting Continuous B-Cell Epitopes. Protein Eng. Des. Sel. 2009, 22, 113–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting Linear B-Cell Epitopes Using String Kernels. J. Mol. Recognit. 2008, 21, 243–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, B.; Zhang, L.; Liang, S.; Zhang, C. SVMTriP: A Method to Predict Antigenic Epitopes Using Support Vector Machine to Integrate Tri-Peptide Similarity and Propensity. PLoS ONE 2012, 7, e45152. [Google Scholar] [CrossRef] [Green Version]
- Singh, H.; Ansari, H.R.; Raghava, G.P.S. Improved Method for Linear B-Cell Epitope Prediction Using Antigen’s Primary Sequence. PLoS ONE 2013, 8, e62216. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, V.; Gautham, N. Harnessing Computational Biology for Exact Linear B-Cell Epitope Prediction: A Novel Amino Acid Composition-Based Feature Descriptor. OMICS A J. Integr. Biol. 2015, 19, 648–658. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune Epitope Database—Analysis Resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Faraggi, E.; Zhou, Y.; Ruan, J.; Kurgan, L. BEST: Improved Prediction of B-Cell Epitopes from Antigen Sequences. PLoS ONE 2012, 7, e40104. [Google Scholar] [CrossRef] [Green Version]
- Tarang, S.; Kesherwani, V.; LaTendresse, B.; Lindgren, L.; Rocha-Sanchez, S.M.; Weston, M.D. In Silico Design of a Multivalent Vaccine Against Candida Albicans. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, S.; Parmar, J.; Kumar, A. Structure-Based Immunogenicity Prediction of Uricase from Fungal (Aspergillus flavus), Bacterial (Bacillus subtillis) and Mammalian Sources Using Immunoinformatic Approach. Protein J. 2020, 39, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and in Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.C. Prediction of Linear B-Cell Epitopes Using Amino Acid Pair Antigenicity Scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking. PLoS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, Z.; Zhang, L.; Yan, D.; Mao, T.; Tang, K.; Qiu, T.; Cao, Z. SEPPA 3.0—Enhanced Spatial Epitope Prediction Enabling Glycoprotein Antigens. Nucleic Acids Res. 2019, 47, W388–W394. [Google Scholar] [CrossRef]
- Sweredoski, M.J.; Baldi, P. PEPITO: Improved Discontinuous B-Cell Epitope Prediction Using Multiple Distance Thresholds and Half Sphere Exposure. Bioinformatics 2008, 24, 1459–1460. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, N.D.; Mayrose, I.; Martz, E.; Pupko, T. Epitopia: A Web-Server for Predicting B-Cell Epitopes. BMC Bioinform. 2009, 10, 287. [Google Scholar] [CrossRef] [Green Version]
- Haste Andersen, P.; Nielsen, M.; Lund, O. Prediction of Residues in Discontinuous B-Cell Epitopes Using Protein 3D Structures. Protein Sci. 2006, 15, 2558–2567. [Google Scholar] [CrossRef]
- Xu, X.L.; Sun, J.; Liu, Q.; Wang, X.J.; Xu, T.L.; Zhu, R.X.; Wu, D.; Cao, Z.W. Evaluation of Spatial Epitope Computational Tools Based on Experimentally-Confirmed Dataset for Protein Antigens. Chin. Sci. Bull. 2010, 55, 2169–2174. [Google Scholar] [CrossRef]
- El-Manzalawy, Y.; Dobbs, D.; Honavar, V.G. In Silico Prediction of Linear B-Cell Epitopes on Proteins. Methods Mol. Biol. 2017, 1484, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.; Ansari, F.A.; Raghunandanan, M.V.; Ramachandran, S. FungalRV: Adhesin Prediction and Immunoinformatics Portal for Human Fungal Pathogens. BMC Genom. 2011, 12, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, R.; Ramachandran, S. Immunoinformatics as a Tool for New Antifungal Vaccines. In Vaccines for Invasive Fungal Infections; Humana Press: New York, NY, USA, 2017; Volume 1625, pp. 31–43. [Google Scholar]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Vivona, S.; Bernante, F.; Filippini, F. NERVE: New Enhanced Reverse Vaccinology Environment. BMC Biotechnol. 2006, 6, 35. [Google Scholar] [CrossRef]
- Xiang, Z.; He, Y. Vaxign: A Web-Based Vaccine Target Design Program for Reverse Vaccinology. Procedia Vaccinol. 2009, 1, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-Throughput Prediction of Protein Antigenicity Using Protein Microarray Data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef]
- Jaiswal, V.; Chanumolu, S.K.; Gupta, A.; Chauhan, R.S.; Rout, C. Jenner-Predict Server: Prediction of Protein Vaccine Candidates (PVCs) in Bacteria Based on Host-Pathogen Interactions. BMC Bioinform. 2013, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Moise, L.; Gutierrez, A.; Kibria, F.; Martin, R.; Tassone, R.; Liu, R.; Terry, F.; Martin, B.; De Groot, A.S. Ivax: An Integrated Toolkit for the Selection and Optimization of Antigens and the Design of Epitope-Driven Vaccines. Hum. Vaccines Immunother. 2015, 11, 2312–2321. [Google Scholar] [CrossRef]
- Rizwan, M.; Naz, A.; Ahmad, J.; Naz, K.; Obaid, A.; Parveen, T.; Ahsan, M.; Ali, A. VacSol: A High Throughput in Silico Pipeline to Predict Potential Therapeutic Targets in Prokaryotic Pathogens Using Subtractive Reverse Vaccinology. BMC Bioinform. 2017, 18, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Doytchinova, I.; Flower, D.R. Bioinformatic Approach for Identifying Parasite and Fungal Candidate Subunit Vaccines. Open Vaccine J. 2008, 1, 4. [Google Scholar] [CrossRef]
- Flower, D.R.; Doytchinova, I.; Zaharieva, N.; Dimitrov, I. Immunogenicity Prediction by VaxiJen: A Ten Year Overview. J. Proteom. Bioinform. 2017, 10, 298–310. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; Von Heijne, G.; Brunak, S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 73. [Google Scholar] [CrossRef]
- Nielsen, H.; Engelbrecht, J. Identification of Prokaryotic and Eukaryotic Signal Peptides and Prediction of Their Cleavage Sites Artificial Neural Networks Have Been Used for Many Biological. Protein Eng. 1997, 10, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of Combined Transmembrane Topology and Signal Peptide Prediction-the Phobius Web Server. Nucleic Acids Res. 2007, 35, 429–432. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting Subcellular Localization of Proteins Based on Their N-Terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melhem, H.; Min, X.J.; Butler, G. The Impact of SignalP 4.0 on the Prediction of Secreted Proteins. In Proceedings of the 2013 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Singapore, 16–19 April 2013; pp. 16–22. [Google Scholar]
- Blum, T.; Briesemeister, S.; Kohlbacher, O. MultiLoc2: Integrating Phylogeny and Gene Ontology Terms Improves Subcellular Protein Localization Prediction. BMC Bioinform. 2009, 10, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabouille, C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017, 27, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Pompa, A.; De Marchis, F.; Pallotta, M.T.; Benitez-Alfonso, Y.; Jones, A.; Schipper, K.; Moreau, K.; Žárský, V.; Di Sansebastiano, G.P.; Bellucci, M. Unconventional Transport Routes of Soluble and Membrane Proteins and Their Role in Developmental Biology. Int. J. Mol. Sci. 2017, 18, 703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.P.; Longo, L.V.G.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and Vesicle-Free Extracellular Proteome of Paracoccidioides brasiliensis: Comparative Analysis with Other Pathogenic Fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef] [Green Version]
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular Transport in Histoplasma capsulatum: An Effective Mechanism for Trans-Cell Wall Transfer of Proteins and Lipids in Ascomycetes. Cell. Microbiol. 2008, 10, 1695–1710. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular Vesicles Produced by Cryptococcus neoformans Contain Protein Components Associated with Virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Sorgo, A.G.; Heilmann, C.J.; Brul, S.; de Koster, C.G.; Klis, F.M. Beyond the Wall: Candida Albicans Secret(e)s to Survive. FEMS Microbiol. Lett. 2013, 338, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Gil-Bona, A.; Llama-Palacios, A.; Parra, C.M.; Vivanco, F.; Nombela, C.; Monteoliva, L.; Gil, C. Proteomics Unravels Extracellular Vesicles as Carriers of Classical Cytoplasmic Proteins in Candida Albicans. J. Proteome Res. 2015, 14, 142–153. [Google Scholar] [CrossRef]
- Souza, J.A.M.; Baltazar, L.D.M.; Carregal, V.M.; Gouveia-Eufrasio, L.; de Oliveira, A.G.; Dias, W.G.; Campos Rocha, M.; Rocha de Miranda, K.; Malavazi, I.; Santos, D.D.A.; et al. Characterization of Aspergillus fumigatus Extracellular Vesicles and Their Effects on Macrophages and Neutrophils Functions. Front. Microbiol. 2019, 10, 2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.S.; et al. Extracellular Vesicles Secreted by Saccharomyces Cerevisiae Are Involved in Cell Wall Remodelling. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-Based Prediction of Non-Classical and Leaderless Protein Secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Poschmann, G.; Waldera-Lupa, D.; Rafiee, N.; Kollmann, M.; Stühler, K. OutCyte: A Novel Tool for Predicting Unconventional Protein Secretion. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Kandaswamy, K.K.; Pugalenthi, G.; Hartmann, E.; Kalies, K.U.; Möller, S.; Suganthan, P.N.; Martinetz, T. SPRED: A Machine Learning Approach for the Identification of Classical and Non-Classical Secretory Proteins in Mammalian Genomes. Biochem. Biophys. Res. Commun. 2010, 391, 1306–1311. [Google Scholar] [CrossRef]
- Ijaq, J.; Chandrasekharan, M.; Poddar, R.; Bethi, N.; Sundararajan, V.S. Annotation and Curation of Uncharacterized Proteins- Challenges. Front. Genet. 2015, 6, 119. [Google Scholar] [CrossRef] [Green Version]
- Aciole Barbosa, D.; Menegidio, F.B.; Alencar, V.C.; Gonçalves, R.S.; de Fatima Santos Silva, J.; Vilas Boas, R.O.; Faustino de Maria, Y.N.L.; Jabes, D.L.; Costa de Oliveira, R.; Nunes, L.R. ParaDB: A Manually Curated Database Containing Genomic Annotation for the Human Pathogenic Fungi Paracoccidioides spp. PLoS Negl. Trop. Dis. 2019, 13, e0007576. [Google Scholar] [CrossRef] [Green Version]
- Silva, P.F.F.; Novaes, E.; Pereira, M.; Soares, C.M.A.; Borges, C.L.; Salem-Izacc, S.M. In Silico Characterization of Hypothetical Proteins from Paracoccidioides lutzii. Genet. Mol. Res. 2015, 14, 17416–17425. [Google Scholar] [CrossRef]
- Gazi, M.A.; Kibria, M.G.; Mahfuz, M.; Islam, M.R.; Ghosh, P.; Afsar, M.N.A.; Khan, M.A.; Ahmed, T. Functional, Structural and Epitopic Prediction of Hypothetical Proteins of Mycobacterium Tuberculosis H37Rv: An in Silico Approach for Prioritizing the Targets. Gene 2016, 591, 442–455. [Google Scholar] [CrossRef]
- Kumar, K.; Prakash, A.; Anjum, F.; Islam, A.; Ahmad, F.; Hassan, M.I. Structure-Based Functional Annotation of Hypothetical Proteins from Candida Dubliniensis: A Quest for Potential Drug Targets. 3 Biotech 2015, 5, 561–576. [Google Scholar] [CrossRef] [Green Version]
- Zambuzzi-Carvalho, P.F.; Fernandes, A.G.; Valadares, M.C.; Tavares, P.D.M.; Nosanchuk, J.D.; de Almeida Soares, C.M.; Pereira, M. Transcriptional Profile of the Human Pathogenic Fungus Paracoccidioides lutzii in Response to Sulfamethoxazole. Med. Mycol. 2015, 53, 477–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, J.; Kavanagh, K. Proteomic Analysis of Proteins Released from Growth-Arrested Candida Albicans Following Exposure to Caspofungin. Med. Mycol. 2010, 48, 598–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihara, D. Protein Function Prediction (Article of SUPERFOCUS); Humana Press: Totowa, NJ, USA, 2017; ISBN 9781493970131. [Google Scholar]
- Lee, D.; Redfern, O.; Orengo, C. Predicting Protein Function from Sequence and Structure. Nat. Rev. Mol. Cell Biol. 2007, 8, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Uddin, R.; Rafi, S. Structural and Functional Characterization of a Unique Hypothetical Protein (WP_003901628.1) of Mycobacterium Tuberculosis: A Computational Approach. Med. Chem. Res. 2017, 26, 1029–1041. [Google Scholar] [CrossRef]
- Bharat Siva Varma, P.; Adimulam, Y.B.; Kodukula, S. In Silico Functional Annotation of a Hypothetical Protein from Staphylococcus Aureus. J. Infect. Public Health 2015, 8, 526–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Zeng, X.; Tsui, S.K.W. Investigating Function Roles of Hypothetical Proteins Encoded by the Mycobacterium Tuberculosis H37Rv Genome. BMC Genom. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivashankari, S.; Shanmughavel, P. Functional Annotation of Hypothetical Proteins—A Review. Bioinformation 2006, 1, 335–338. [Google Scholar] [CrossRef] [Green Version]
- Mills, C.L.; Beuning, P.J.; Ondrechen, M.J. Biochemical Functional Predictions for Protein Structures of Unknown or Uncertain Function. Comput. Struct. Biotechnol. J. 2015, 13, 182–191. [Google Scholar] [CrossRef] [Green Version]
- Tatibana, B.T.; Sano, A.; Uno, J.; Kamei, K.; Igarashi, T.; Mikami, Y.; Miyaji, M.; Nishimura, K.; Itano, E.N. Detection of Paracoccidioides brasiliensis Gp43 Gene in Sputa by Loop-Mediated Isothermal Amplification Method. J. Clin. Lab. Anal. 2009, 23, 139–143. [Google Scholar] [CrossRef]
- Endo, S.; Komori, T.; Ricci, G.; Sano, A.; Yokoyama, K.; Ohori, A.; Kamei, K.; Franco, M.; Miyaji, M.; Nishimura, K. Detection of Gp43 of Paracoccidioides brasiliensis by the Loop-Mediated Isothermal Amplification (LAMP) Method. FEMS Microbiol. Lett. 2004, 234, 93–97. [Google Scholar] [CrossRef]
- Carvajal, D.L.M. Desenvolvimento de Método Diagnóstico Através do Desenho de Sondas Para Identificação de Paracoccidioides lutzii, Utilizando a Técnica de Loop—Mediated Isothermal Amplification (LAMP); Faculdade de Ciências Médicas, Universidade Estadual de Campinas: Campinas, Brazil, 2018. [Google Scholar]
- Rosa, S.B.A.; Csordas, B.G.; do Valle Leone de Oliveira, S.M.; Ribeiro dos Santos, A.; Paniago, A.M.M.; Venturini, J. Prediction of Conserved Peptides of Paracoccidioides for Interferon-γ Release Assay: The First Step in the Development of a Lab-Based Approach for Immunological Assessment during Antifungal Therapy. J. Fungi 2020, 6, 379. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, D.H.; Gómez, B.L.; Tobón, A.M.; Chiller, T.M.; Lindsley, M.D. Evaluation of a Histoplasma Antigen Lateral Flow Assay for the Rapid Diagnosis of Progressive Disseminated Histoplasmosis in Colombian Patients with AIDS. Mycoses 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Linder, K.A.; Kauffman, C.A. Histoplasmosis: Epidemiology, Diagnosis, and Clinical Manifestations. Curr. Fungal Infect. Rep. 2019, 13, 120–128. [Google Scholar] [CrossRef]
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.A.; Colombo, A.L.; Hoenigl, M.; Klimko, N.; Lass-Flörl, C.; Oladele, R.O.; Vinh, D.C.; et al. Defining and Managing COVID-19-Associated Pulmonary Aspergillosis: The 2020 ECMM/ISHAM Consensus Criteria for Research and Clinical Guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef]
- Permpalung, N.; Maertens, J.; Marr, K.A. Diagnostic Dilemma in COVID-19-Associated Pulmonary Aspergillosis. Lancet Infect. Dis. 2021, 21, 766–767. [Google Scholar] [CrossRef]
- Arastehfar, A.; Carvalho, A.; Van De Veerdonk, F.L.; Jenks, J.D.; Koehler, P.; Krause, R.; Cornely, O.A.; Perlin, D.S.; Lass-Flörl, C.; Hoenigl, M. COVID-19 Associated Pulmonary Aspergillosis (CAPA)—From Immunology to Treatment. J. Fungi 2020, 6, 91. [Google Scholar] [CrossRef]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inácio, M.M.; Cruz-Leite, V.R.M.; Moreira, A.L.E.; Mattos, K.; Paccez, J.D.; Ruiz, O.H.; Venturini, J.; de Souza Carvalho Melhem, M.; Paniago, A.M.M.; de Almeida Soares, C.M.; et al. Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets. Pathogens 2022, 11, 569. https://doi.org/10.3390/pathogens11050569
Inácio MM, Cruz-Leite VRM, Moreira ALE, Mattos K, Paccez JD, Ruiz OH, Venturini J, de Souza Carvalho Melhem M, Paniago AMM, de Almeida Soares CM, et al. Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets. Pathogens. 2022; 11(5):569. https://doi.org/10.3390/pathogens11050569
Chicago/Turabian StyleInácio, Moisés Morais, Vanessa Rafaela Milhomem Cruz-Leite, André Luís Elias Moreira, Karine Mattos, Juliano Domiraci Paccez, Orville Hernandez Ruiz, James Venturini, Marcia de Souza Carvalho Melhem, Anamaria Mello Miranda Paniago, Célia Maria de Almeida Soares, and et al. 2022. "Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets" Pathogens 11, no. 5: 569. https://doi.org/10.3390/pathogens11050569
APA StyleInácio, M. M., Cruz-Leite, V. R. M., Moreira, A. L. E., Mattos, K., Paccez, J. D., Ruiz, O. H., Venturini, J., de Souza Carvalho Melhem, M., Paniago, A. M. M., de Almeida Soares, C. M., Weber, S. S., & Borges, C. L. (2022). Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets. Pathogens, 11(5), 569. https://doi.org/10.3390/pathogens11050569