Dynamics of Infections in Cattle and Rhipicephalus microplus: A Preliminary Study
Abstract
:1. Introduction
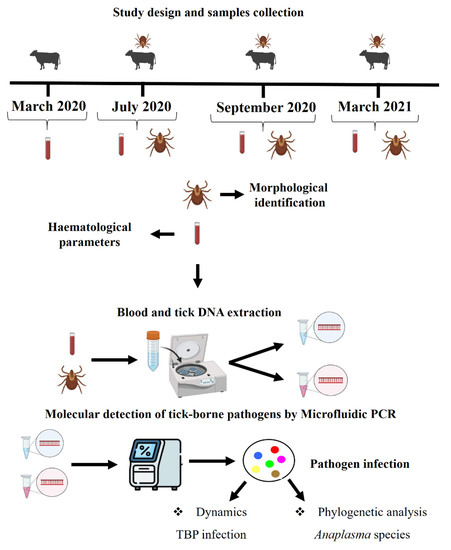
2. Materials and Methods
2.1. Study Design
2.2. Blood Samples Collection and Haematological Parameters
2.3. Tick Collection and Morphological Identification
2.4. Blood and Tick DNA Extraction
2.5. Molecular Detection of Tick-Borne Pathogens
2.5.1. DNA Pre-Amplification for Real-Time Microfluidic PCR
2.5.2. Real-Time Microfluidic PCR
2.5.3. PCR and DNA Sequencing for Anaplasma Species Identification
2.6. Phylogenetic Analysis
3. Results
3.1. Bovine Hematological Parameters and Taxonomic Identification of Collected Ticks
3.2. Detection of TBPs in Cattle Samples and Ticks
3.3. Dynamics of Tick-Borne Pathogen Infection in Cattle and Ticks
3.4. Phylogenetic Analysis of Identified Anaplasma Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kolte, S.W.; Larcombe, S.D.; Jadhao, S.G.; Magar, S.P.; Warthi, G.; Kurkure, N.V.; Glass, E.J.; Shiels, B.R. PCR diagnosis of tick-borne pathogens in Maharashtra state, India indicates fitness cost associated with carrier infections is greater for crossbreed than native cattle breeds. PLoS ONE 2017, 12, 0174595. [Google Scholar] [CrossRef] [Green Version]
- Satti, R.A.; Awadelkareem, E.A.; Suganuma, K.; Salim, B.; Inoue, N.; Xuan, X.; Rehan, S.; Mossaad, E. Cattle anaplasmosis and babesiosis: Major tick-borne diseases affecting the cattle industry in Khartoum State, Sudan. Vet. Parasitol. Reg. Stud. Rep. 2021, 26, 100632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, K.; Sun, Y.; Shi, J.; Li, H.; Chen, Y.; Yang, H.; Li, X.; Wu, B.; Li, X.; et al. Molecular epidemiology and risk factors of Anaplasma spp., Babesia spp. and Theileria spp. infection in cattle in Chongqing, China. PLoS ONE 2019, 14, 0215585. [Google Scholar] [CrossRef] [Green Version]
- Bonilla-Aldana, D.K.; Quintero-Rada, K.; Montoya-Posada, J.P.; Soler-Tovar, D.; Barato, P.; Arteaga-Livias, K.; Zambrano, L.I.; Faccini-Martínez, Á.A.; Rodriguez-Morales, A.J. Bovine Ehrlichiosis Prevalence: A Systematic Review and Meta-Analysis of Molecular Studies. World Vet. J. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Suarez, C.E.; Noh, S. Emerging perspectives in the research of bovine babesiosis and anaplasmosis. Vet. Parasitol. 2011, 180, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Obregón Alvarez, D.; Corona-González, B.; Rodríguez-Mallón, A.; Rodríguez Gonzalez, I.; Alfonso, P.; Noda Ramos, A.A.; Díaz-Sánchez, A.A.; González Navarrete, M.; Rodríguez Fernández, R.; Méndez Mellor, L.; et al. Ticks and tick-borne diseases in Cuba, half a century of scientific research. Pathogens 2020, 9, 616. [Google Scholar] [CrossRef]
- Obregón, D.; Cabezas-Cruz, A.; Armas, Y.; Silva, J.B.; Fonseca, A.H.; André, M.R.; Alfonso, P.; Oliveira, M.C.; Machado, R.Z.; Corona-González, B. High co-infection rates of Babesia bovis, Babesia bigemina, and Anaplasma marginale in water buffalo in Western Cuba. Parasitol. Res. 2019, 118, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Aubry, P.; Dorothy, W.G. A review of bovine anaplasmosis. Transbound. Emerg. Dis. 2011, 58, 1–30. [Google Scholar] [CrossRef]
- Salinas-Estrella, E.; Amaro-Estrada, I.; Cobaxin-Cárdenas, M.E.; Preciado de la Torre, J.F.; Rodríguez, S.D. Bovine Anaplasmosis: Will there ever be an almighty effective vaccine? Front. Vet. Sci. 2022, 9, 946545. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, N.N.; Bock, R.E.; Jorgensen, W.K. Productivity and health effects of anaplasmosis and babesiosis on Bos indicus cattle and their crosses, and the effects of differing intensity of tick control in Australia. Vet. Parasitol. 2008, 155, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Sequeira, T.C.G.; Oliveira, M.D.S.; Araujo, J.P., Jr.; Amarante, A.F.T. PCR-based detection of Babesia bovis and Babesia bigemina in their natural host Boophilus microplus and cattle. Int. J. Parasitol. 2005, 35, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Reetha, T.L.; Thomas, K.S.; Babu, M. Occurrence of haemoprotozoan infection in bovines. Int. J. Appl. Biores 2012, 13, 1–2. [Google Scholar]
- Maharana, B.R.; Tewari, A.K.; Saravanan, B.C.; Sudhakar, N.R. Important hemoprotozoan diseases of livestock: Challenges in current diagnostics and therapeutics: An update. Vet. World 2016, 9, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.B.; Alagib, A.; AbdElkareim, T.B.; Hassan, M.M.; Johnson, W.C.; Hussein, H.E.; Taus, N.S.; Ueti, M.W. Molecular detection and characterization of Theileria spp. infecting cattle in Sennar State, Sudan. Parasitol. Res. 2018, 117, 1271–1276. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, L.; Bouda, J. Patología clínica Veterinaria; UNAM, Facultad de medicina veterinaria y zootecnia: Mexico City, Mexico, 2007. [Google Scholar]
- Jain, N.C. Essentials of Veterinary Hematology; Lea & Febiger: Philadelphia, PA, USA, 1993; p. 417. [Google Scholar]
- Meyer, D.J.; Harvey, J.W. Veterinary Laboratory Medicine: Interpretation & Diagnosis, 2nd ed.; Sauders: Philadelphia, PA, USA, 2004; p. 351. [Google Scholar]
- Alberghina, D.; Giannetto, C.; Vazzana, I.; Ferrantelli, V.; Piccione, G. Reference intervals for total protein concentration, serum protein fractions, and albumin/globulin ratios in clinically healthy dairy cows. J. Vet. Diagn. Investig. 2011, 23, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Estrada-Peña, A.; Bouattour, A.; Camicas, J.; Walker, A.R. Tick of Domestic Animals in Mediterranean Region: A Guide to Identification of Species; University of Zaragoza Press: Zaragoza, Spain, 2004; pp. 130–133. [Google Scholar]
- Gondard, M.; Delannoy, S.; Pinarello, V.; Aprelon, R.; Devillers, E.; Galon, C.; Pradel, J.; Vayssier-Taussat, M.; Albina, E.; Moutailler, S. Upscaling the surveillance of tick-borne pathogens in the French Caribbean islands. Pathogens 2020, 9, 176. [Google Scholar] [CrossRef] [Green Version]
- Grech-Angelini, S.; Stachurski, F.; Vayssier-Taussat, M.; Devillers, E.; Casabianca, F.; Lancelot, R.; Uilenberg, G.; Moutailler, S. Tick-borne pathogens in ticks (Acari: Ixodidae) collected from various domestic and wild hosts in Corsica (France), a Mediterranean island environment. Transbound. Emerg. Dis. 2020, 67, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Delannoy, S.; Devillers, E.; Umhang, G.; Aspan, A.; Juremalm, M.; Chirico, J.; van der Wal, F.J.; Sprong, H.; Pihl, T.P.B.; et al. High-throughput screening of tick-borne pathogens in Europe. Front. Cell Infect. Microbiol. 2014, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Rar, V.A.; Fomenko, N.V.; Dobrotvorsky, A.K.; Livanova, N.N.; Rudakova, S.A.; Fedorov, E.G.; Astanin, V.B.; Morozova, O.V. Tick-borne pathogen detection, Western Siberia, Russia. Emerg. Infect. Dis. 2005, 11, 1708–1715. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Etim, N.N.; Williams, M.E.; Akpabio, U.; Offiong, E.E. Haematological parameters and factors affecting their values. Agric. Sci. 2014, 2, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Onasanya, G.O.; Oke, F.O.; Sanni, T.M.; Muhammad, A.I. Parameters influencing haematological, serum and bio-chemical references in livestock animals under different management systems. Open J. Vet. Med. 2015, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Martins, K.R.; Garcia, M.V.; Bonatte-Junior, P.; Duarte, P.O.; de Higa, L.O.S.; Csordas, B.G.; Barros, J.C.; Andreotti, R. Correlation between Rhipicephalus microplus ticks and Anaplasma marginale infection in various cattle breeds in Brazil. Exp. Appl. Acarol. 2020, 81, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Haque, M.; Singh, N.K.; Rath, S.S. Molecular detection of Anaplasma marginale infection in carrier cattle. Ticks Tick-Borne Dis. 2012, 3, 55–58. [Google Scholar] [CrossRef]
- Noaman, V.; Shayan, P. Molecular detection of Anaplasma phagocytophilum in carrier cattle of Iran-first documented report. Iran. J. Microbiol. 2009, 1, 37–42. [Google Scholar]
- Nair, A.S.; Ravindran, R.; Lakshmanan, B.; Sreekumar, C.; Kumar, S.S.; Raju, R.; Tresamol, P.V.; Vimalkumar, M.B.; Saseendranath, M.R. Bovine carriers of Anaplasma marginale and Anaplasma bovis in South India. Trop. Biomed. 2013, 30, 105–112. [Google Scholar]
- Ghafar, A.; Davies, N.; Tadepalli, M.; Breidahl, A.; Death, C.; Haros, P.; Li, Y.; Dann, P.; Cabezas-Cruz, A.; Moutailler, S.; et al. Unravelling the Diversity of Microorganisms in Ticks from Australian Wildlife. Pathogens 2023, 12, 153. [Google Scholar] [CrossRef]
- Guizzo, M.G.; Parizi, L.F.; Nunes, R.D.; Schama, R.; Albano, R.M.; Tirloni, L.; Oldiges, D.P.; Vieira, R.P.; Oliveira, W.H.; Leite, M.D. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci. Rep. 2017, 7, 17554. [Google Scholar] [CrossRef] [Green Version]
- Kieser, S.T.; Eriks, I.S.; Palmer, G.H. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect. Immun. 1990, 58, 1117–1119. [Google Scholar] [CrossRef]
- Lohr, C.V.; Brayton, K.A.; Shkap, V.; Molad, T.; Barbet, A.F.; Brown, W.C.; Palmer, G.H. Expression of Anaplasma marginale major surface protein 2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 2002, 70, 6005–6012. [Google Scholar] [CrossRef] [Green Version]
- Eriks, I.S.; Stiller, D.; Palmer, G.H. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J. Clin. Microbiol. 1993, 31, 2091–2096. [Google Scholar] [CrossRef]
- Kalil, S.P.; Rosa, R.D.; Capelli-Peixoto, J.; Pohl, P.C.; Oliveira, P.L.; Fogaça, A.C.; Daffre, S. Immune-related redox metabolism of embryonic cells of the tick Rhipicephalus microplus (BME26) in response to infection with Anaplasma marginale. Parasites Vectors 2017, 10, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenneil, R.; Shkap, V.; Leibovich, B.; Zweygarth, E.; Pfister, K.; Ribeiro, M.F.; Passos, L.M. Cross-Protection Between Geographically Distinct Anaplasma marginale Isolates Appears to be Constrained by Limited Antibody Responses. Transbound. Emerg. Dis. 2013, 60, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kocan, K.M.; de la Fuente, J.; Guglielmone, A.A.; Melendez, R.D. Antigens and alternatives for control ofAnaplasma marginaleinfection in cattle. Clin. Microbiol. Rev. 2003, 16, 698–712. [Google Scholar] [CrossRef] [Green Version]
- Kocan, K.M.; De La Fuente, J.; Blouin, E.F.; Garcia-Garcia, J.C. Anaplasma marginale (Rickettsiales: Anaplasmataceae): Recent advances in defining host–pathogen adaptations of a tick-borne rickettsia. Parasitology 2004, 129, S285–S300. [Google Scholar] [CrossRef] [PubMed]
- Belkahia, H.; Ben Said, M.; Alberti, A.; Abdi, K.; Issaoui, Z.; Hattab, D.; Gharbi, M.; Messadi, L. First molecular survey and novel genetic variants’ identification of Anaplasma marginale, A. centrale and A. bovis in cattle from Tunisia. Infect. Genet. Evol. 2015, 34, 361–371. [Google Scholar] [CrossRef]
- Belkahia, H.; Ben Said, M.; El Mabrouk, N.; Saidani, M.; Cherni, C.; Ben Hassen, M.; Bouattour, A.; Messadi, L. Spatio-temporal variations and genetic diversity of Anaplasma spp. in cattle from the North of Tunisia. Vet. Microbiol. 2017, 208, 223–230. [Google Scholar] [CrossRef]
- M’ghirbi, Y.; Bèji, M.; Oporto, B.; Khrouf, F.; Hurtado, A.; Bouattour, A. Anaplasma marginale and A. phagocytophilum in cattle in Tunisia. Parasites Vectors 2016, 9, 556. [Google Scholar] [CrossRef] [Green Version]
- Bilgiç, H.B.; Karagenç, T.; Simuunza, M.; Shiels, B.; Tait, A.; Eren, H.; Weir, W. Development of a multiplex PCR assay for simultaneous detection of Theileria annulata, Babesia bovis and Anaplasma marginale in cattle. Exp. Parasitol. 2013, 133, 222–229. [Google Scholar] [CrossRef] [Green Version]
- Oakes, V.J.; Todd, S.M.; Carbonello, A.A.; Michalak, P.; Lahmers, K.K. Coinfection of cattle in Virginia with Theileria orientalis Ikeda genotype and Anaplasma marginale. J. Vet. Diagn. Investig. 2022, 34, 36–41. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Thumbi, S.M.; Jennings, A.; Chase-Topping, M.; Callaby, R.; Kiara, H.; Oosthuizen, M.C.; Mbole-Kariuki, M.N.; Conradie, I.; Handel, I.G.; et al. Co-infections determine patterns of mortality in a population exposed to parasite infection. Sci. Adv. 2015, 1, 1400026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatanga, E.; Maganga, E.; Mohamed, W.M.A.; Ogata, S.; Pandey, G.S.; Abdelbaset, A.E.; Hayashida, K.; Sugimoto, C.; Katakura, K.; Nonaka, N.; et al. High infection rate of tick-borne protozoan and rickettsial pathogens of cattle in Malawi and the development of a multiplex PCR for Babesia and Theileria species identification. Acta Trop. 2022, 231, 106413. [Google Scholar] [CrossRef] [PubMed]
- Reif, K.E.; Macaluso, K.R. Ecology of Rickettsia felis: A review. J. Med. Entomol. 2009, 46, 723–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirunkanokpun, S.; Thepparit, C.; Foil, L.D.; Macaluso, K.R. Horizontal transmission of Rickettsia felis between cat fleas, Ctenocephalides felis. Mol. Ecol. 2011, 20, 4577–4586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazid Abdad, M.; Stenos, J.; Graves, S. Rickettsia felis, an emerging flea-transmitted human pathogen. Emerg. Health Threat. J. 2011, 4, 7168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abanda, B.; Paguem, A.; Abdoulmoumini, M.; Kingsley, M.T.; Renz, A.; Eisenbarth, A. Molecular identification and prevalence of tick-borne pathogens in zebu and taurine cattle in North Cameroon. Parasites Vectors 2019, 12, 448. [Google Scholar] [CrossRef] [Green Version]
- Danis-Lozano, R.; Camacho-Ramírez, S.; Álvarez-Hernández, G.; Leyva-Gastelum, M.; Cisneros-Vásquez, L.A.; Dzul-Rosado, K.R.; Fernández-Salas, I.; López-Ordoñez, T. Evidencia molecular de Rickettsia ricketsii y Rickettsia felis en garrapatas colectadas en ganado bovino en la costa de Chiapas. Salud Pública México 2023, 65, 160–166. [Google Scholar] [CrossRef]
- Díaz-Sánchez, A.A.; Chilton, N.B.; Roblejo-Arias, L.; Fonseca-Rodríguez, O.; Marrero-Perera, R.; Diyes, C.P.; Yunik, M.E.; Lobo-Rivero, E.; Corona-González, B. Molecular detection and identification of spotted fever group rickettsiae in ticks collected from horses in Cuba. Med. Vet. Entomol. 2021, 35, 207–212. [Google Scholar] [CrossRef]
- Dyachenko, V.; Pantchev, N.; Balzer, H.J.; Meyersen, A.; Straubinger, R.K. First case of Anaplasma platys infection in a dog from Croatia. Parasites Vectors 2012, 5, 49. [Google Scholar] [CrossRef] [Green Version]
- Bouzouraa, T.; René-Martellet, M.; Chêne, J.; Attipa, C.; Lebert, I.; Chalvet-Monfray, K.; Cadoré, J.L.; Halos, L.; Chabanne, L. Clinical and laboratory features of canine Anaplasma platys infection in 32 naturally infected dogs in the Mediterranean basin. Ticks Tick-Borne Dis. 2016, 7, 1256–1264. [Google Scholar] [CrossRef]
- Snellgrove, A.N.; Krapiunaya, I.; Ford, S.L.; Stanley, H.M.; Wickson, A.G.; Hartzer, K.L.; Levin, M.L. Vector competence of Rhipicephalus sanguineus sensu stricto for Anaplasma platys. Ticks Tick-Borne Dis. 2020, 11, 101517. [Google Scholar] [CrossRef] [PubMed]
- Chien, N.T.H.; Nguyen, T.L.; Bui, K.L.; Van Nguyen, T.; Le, T.H. Anaplasma marginale and A. platys characterized from dairy and indigenous cattle and dogs in northern Vietnam. Korean J. Parasitol. 2019, 57, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahmani, M.; Davoust, B.; Benterki, M.S.; Fenollar, F.; Raoult, D.; Mediannikov, O. Development of a new PCR-based assay to detect Anaplasmataceae and the first report of Anaplasma phagocytophilum and Anaplasma platys in cattle from Algeria. Comp. Immunol. Microbiol. Infect. Dis. 2015, 39, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Tumwebaze, M.A.; Lee, S.H.; Moumouni, P.F.A.; Mohammed-Geba, K.; Sheir, S.K.; Galal-Khallaf, A.; Abd El Latif, H.M.; Morsi, D.S.; Boshr, N.M.; Galon, E.M.; et al. First detection of Anaplasma ovis in sheep and Anaplasma platys-like variants from cattle in Menoufia governorate, Egypt. Parasitol. Int. 2020, 78, 102150. [Google Scholar] [CrossRef] [PubMed]
- Kamani, J.; Schaer, J.; Umar, A.G.; Pilarshimwi, J.Y.; Bukar, L.; González-Miguel, J.; Harrus, S. Molecular detection and genetic characterization of Anaplasma marginale and Anaplasma platys in cattle in Nigeria. Ticks Tick-Borne Dis. 2022, 13, 101955. [Google Scholar] [CrossRef] [PubMed]
- Corona, B.; Obregón, D.; Martínez, S.; Espinosa, I.; Fonseca, A.; Roque, E. Detección por PCR de Anaplasma marginale en búfalos de la región occidental de Cuba. Rev. Salud Anim. 2012, 34, 11–18. [Google Scholar]
- Díaz-Sánchez, A.A.; Meli, M.L.; Álvarez, D.O.; Fonseca-Rodríguez, O.; Cabezas-Cruz, A.; Hofmann-Lehmann, R.; Corona-González, B. Development and application of a multiplex TaqMan® real-time qPCR assay for the simultaneous detection of Anaplasma marginale and Theileria annulata and molecular characterization of Anaplasma marginale from cattle in Western Cuba. Ticks Tick-Borne Dis. 2020, 11, 101356. [Google Scholar] [CrossRef]
- Corona, B.G.; Díaz-Sánchez, A.A.; Meli, M.L.; Cañizares, E.V.; Arias, L.R.; Dorta, Y.L.; Rivero, E.L.; Hofmann-Lehmann, R. Occurrence of tick-borne pathogens in stray dogs from Havana, Cuba. Acta Biomed. Sci. 2018, 3, 158–159. [Google Scholar] [CrossRef]
- da Silva, C.B.; Santos, H.A.; Navarrete, M.G.; Ribeiro, C.C.; Gonzalez, B.C.; Zaldivar, M.F.; Pires, M.S.; Peckle, M.; da Costa, R.L.; Vitari, G.L.V.; et al. Molecular detection and characterization of Anaplasma platys in dogs and ticks in Cuba. Ticks Tick-Borne Dis. 2016, 7, 938–944. [Google Scholar] [CrossRef]
- Arraga-Alvarado, C.M.; Qurollo, B.A.; Parra, O.C.; Berrueta, M.A.; Hegarty, B.C.; Breitschwerdt, E.B. Case report: Molecular evidence of Anaplasma platys infection in two women from Venezuela. Am. J. Trop. Med. Hy. 2014, 91, 1161–1165. [Google Scholar] [CrossRef]
- National Research Council. The Development of Science-based Guidelines for Laboratory Animal Care: Proceedings of the November 2003 International Workshop; The National Academies Press: Washington, DC, USA, 2004. [Google Scholar] [CrossRef]


| Animals ID | HCT * 0.27–0.47 (L/L) | WBCs * 4.0–12.0 (c/L) | Total Protein Values 56.9–78.7 (g/L) | |||
|---|---|---|---|---|---|---|
| March 2020 | July 2020 | March 2020 | July 2020 | March 2020 | July 2020 | |
| C1 | 0.32 | 0.32 | 22.95 | 11.30 | 82 | 72 |
| C2 | 0.30 | 0.30 | 24.00 | 11.65 | 72 | 62 |
| C3 | 0.30 | 0.30 | 24.40 | 10.25 | 76 | 64 |
| C4 | 0.30 | 0.30 | 7.80 | 8.00 | 74 | 70 |
| C5 | 0.39 | 0.39 | 24.00 | 10.80 | 62 | 58 |
| C6 | 0.32 | 0.32 | 9.95 | 8.05 | 76 | 70 |
| C7 | 0.30 | 0.30 | 12.05 | 11.00 | 58 | 74 |
| C8 | 0.30 | 0.30 | 11.65 | 10.95 | 73 | 68 |
| Vector-Borne Pathogen(s) | Total | % | 95% CI a |
|---|---|---|---|
| Total infected samples (≥1 pathogen) | 32 | 100 | 95–100 |
| A. marginale | 31 | 96.9 | 90.9–100 |
| R. felis | 4 | 12.5 | 1.10–23.9 |
| Rickettsia spp. | 2 | 6.25 | 0–7.09 |
| Anaplasma spp. | 1 | 3.12 | 0–9.19 |
| Single infections | 26 | 81.3 | 67.8–94.8 |
| A. marginale | 26 | 81.3 | 67.8–94.8 |
| Mixed infections | 6 | 18.8 | 5.3–32.5 |
| R. felis + A. marginale | 3 | 9.38 | 0–19.5 |
| A. marginale+ Rickettsia spp. | 2 | 6.25 | 0–7.09 |
| R. felis+ Anaplasma spp. | 1 | 3.13 | 0–0.11 |
| Vector-Borne Pathogen(s) | Total | % | 95% CI a |
|---|---|---|---|
| Total infected ticks (≥1 pathogen) | 15 | 62.5 | 60.6–64.4 |
| A. marginale | 11 | 45.8 | 25.9–65.7 |
| R. felis | 4 | 16.7 | 1.81–31.5 |
| Rickettsia spp. | 1 | 4.17 | 0.1–8.24 |
| Ehrlichia spp. | 2 | 8.33 | 2.69–13.9 |
| Single infections | 12 | 50.0 | 41.4–58.6 |
| A. marginale | 8 | 33.3 | 14.2–51.8 |
| R. felis | 3 | 12.5 | 5.75–19.3 |
| Ehrlichia spp. | 1 | 4.17 | 0.1–8.24 |
| Mixed infections | 3 | 12.5 | 5.75–19.3 |
| A. marginale + Rickettsia spp. | 1 | 4.17 | 0.1–8.24 |
| A. marginale + Rickettsia felis | 1 | 4.17 | 0.1–8.24 |
| B. canis (subspecies) + A. marginale + Ehrlichia spp. | 1 | 4.17 | 0.1–8.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piloto-Sardiñas, E.; Foucault-Simonin, A.; Wu-Chuang, A.; Mateos-Hernández, L.; Marrero-Perera, R.; Abuin-Denis, L.; Roblejo-Arias, L.; Díaz-Corona, C.; Zając, Z.; Kulisz, J.; et al. Dynamics of Infections in Cattle and Rhipicephalus microplus: A Preliminary Study. Pathogens 2023, 12, 998. https://doi.org/10.3390/pathogens12080998
Piloto-Sardiñas E, Foucault-Simonin A, Wu-Chuang A, Mateos-Hernández L, Marrero-Perera R, Abuin-Denis L, Roblejo-Arias L, Díaz-Corona C, Zając Z, Kulisz J, et al. Dynamics of Infections in Cattle and Rhipicephalus microplus: A Preliminary Study. Pathogens. 2023; 12(8):998. https://doi.org/10.3390/pathogens12080998
Chicago/Turabian StylePiloto-Sardiñas, Elianne, Angélique Foucault-Simonin, Alejandra Wu-Chuang, Lourdes Mateos-Hernández, Roxana Marrero-Perera, Lianet Abuin-Denis, Lisset Roblejo-Arias, Cristian Díaz-Corona, Zbigniew Zając, Joanna Kulisz, and et al. 2023. "Dynamics of Infections in Cattle and Rhipicephalus microplus: A Preliminary Study" Pathogens 12, no. 8: 998. https://doi.org/10.3390/pathogens12080998
APA StylePiloto-Sardiñas, E., Foucault-Simonin, A., Wu-Chuang, A., Mateos-Hernández, L., Marrero-Perera, R., Abuin-Denis, L., Roblejo-Arias, L., Díaz-Corona, C., Zając, Z., Kulisz, J., Woźniak, A., Moutailler, S., Corona-González, B., & Cabezas-Cruz, A. (2023). Dynamics of Infections in Cattle and Rhipicephalus microplus: A Preliminary Study. Pathogens, 12(8), 998. https://doi.org/10.3390/pathogens12080998