Isospora and Lankesterella Parasites (Eimeriidae, Apicomplexa) of Passeriform Birds in Europe: Infection Rates, Phylogeny, and Pathogenicity
Abstract
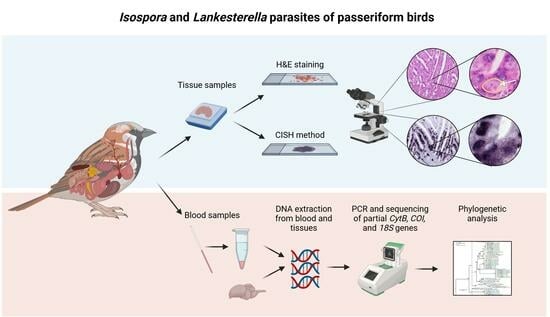
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. Primer Design
2.4. PCR and Sequencing
2.5. Phylogenetic Analysis
2.6. Genetic Distances between Isospora and Lankesterella CytB Lineages
2.7. Chromogenic In Situ Hybridization
2.8. Microscopic Examination
2.9. Graphical Representation of the Parasites’ Life Cycles
3. Results
3.1. Infection Rates
3.2. Phylogeny
3.3. Chromogenic In Situ Hybridization
3.4. Detection of Isospora by CISH
3.5. Detection of Lankesterella by CISH
3.6. Histopathology of Isospora Infections
3.7. Histopathology of Lankesterella Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, J.R.; Lainson, R.; Killick-Kendrick, R. Lankesterella corvi n. sp., a blood parasite of the English rook, Corvus f. frugilegus L. J. Protozool. 1959, 6, 233–238. [Google Scholar] [CrossRef]
- Box, E.D. Influence of Isospora infections on patency of avian Lankesterella (Atoxoplasma, Garnham, 1950). J. Parasitol. 1967, 53, 1140–1147. [Google Scholar] [CrossRef]
- Khan, R.A.; Desser, S.S. Avian Lankesterella infections in Algonquin Park, Ontario. Can. J. Zool. 1971, 49, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Lainson, R. Atoxoplasma Garnham, 1950, as a synonym for Lankesterella Labbé, 1899. Its life cycle in the English sparrow (Passer domesticus domesticus, Linn.). J. Protozool. 1959, 6, 360–371. [Google Scholar] [CrossRef]
- Lainson, R. The transmission of Lankesterella (= Atoxoplasma) in birds by the mite Dermanyssus gallinae. J. Protozool. 1960, 7, 321–322. [Google Scholar] [CrossRef]
- Chagas, C.R.F.; Harl, J.; Preikša, V.; Bukauskaitė, D.; Ilgūnas, M.; Weissenböck, H.; Valkiūnas, G. Lankesterella (Apicomplexa, Lankesterellidae) blood parasites of passeriform birds: Prevalence, molecular and morphological characterization, with notes on sporozoite persistence in vivo and development in vitro. Animals 2021, 11, 1451. [Google Scholar] [CrossRef]
- Biedrzycka, A.; Kloch, A.; Migalska, M.; Bielański, W. Molecular characterization of putative Hepatozoon sp. from the sedge warbler (Acrocephalus schoenobaenus). Parasitology 2013, 140, 695–698. [Google Scholar] [CrossRef]
- Martínez, J.; Merino, S.; Badás, E.P.; Almazán, L.; Moksnes, A.; Barbosa, A. Hemoparasites and immunological parameters in snow bunting (Plectrophenax nivalis) nestlings. Polar Biol. 2018, 41, 1855–1866. [Google Scholar] [CrossRef]
- Merino, S.; Martínez, J.; Martínez-de la Puente, J.; Criado-Fornelio, Á.; Tomás, G.; Morales, J.; Lobato, E.; García-Fraile, S. Molecular characterization of the 18S rDNA gene of an avian Hepatozoon reveals that it is closely related to Lankesterella. J. Parasitol. 2006, 92, 1330–1335. [Google Scholar] [CrossRef]
- Schrenzel, M.D.; Maalouf, G.A.; Gaffney, P.M.; Tokarz, D.; Keener, L.L.; McClure, D.; Griffey, S.; McAloose, D.; Rideout, B.A. Molecular characterization of isosporoid Coccidia (Isospora and Atoxoplasma spp.) in passerine birds. J. Parasitol. 2005, 91, 635–647. [Google Scholar] [CrossRef]
- Barta, J.R.; Schrenzel, M.D.; Carreno, R.; Rideout, B.A. The genus Atoxoplasma (Garnham 1950) as a junior objective synonym of the genus Isospora (Schneider 1881) species infecting birds and resurrection of Cystoisospora (Frenkel 1977) as the correct genus for Isospora species infecting mammals. J. Parasitol. 2005, 91, 726–727. [Google Scholar] [CrossRef] [PubMed]
- Box, E.D. Life cycles of two Isospora species in the canary, Serinus canarius Linnaeus. J. Protozool. 1977, 24, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Flach, E.J.; Dodhia, H.S.; Guthrie, A.; Blake, D.P. Systemic isosporiasis (atoxoplasmosis) in passerine birds at the Zoological Society of London, London Zoo. J. Zoo Wildl. Med. 2022, 53, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Labbé, A. Sur les coccidies des oiseaux. Comptes Rendus l’Académie Sci. 1893, 116, 1300–1303. [Google Scholar]
- Ball, S.J.; Brown, M.A.; Daszak, P.; Pittilo, R.M. Atoxoplasma (Apicomplexa: Eimeriorina: Atoxoplasmatidae) in the greenfinch (Carduelis chloris). J. Parasitol. 1998, 84, 813–817. [Google Scholar] [CrossRef]
- Boulard, Y.; Landau, I.; Grulet, O.; Baccam, D. Ultrastructure of chronic reticuloendothelial forms of Isospora of sparrows. Ann. Parasitol. Hum. Comp. 1987, 62, 181–184. [Google Scholar] [PubMed]
- Greiner, E.C. Isospora, Atoxoplasma, and Sarcocystis. In Parasitic Diseases of Wild Birds; Wiley: Hoboken, NJ, USA, 2008; pp. 108–119. [Google Scholar]
- Terio, K.A.; Adkesson, M.J. Systemic isosporosis in passerine birds. In Fowler’s Zoo and Wild Animal Medicine Current Therapy; Elsevier: Amsterdam, The Netherlands, 2019; Volume 9, pp. 454–458. ISBN 978-0-323-55228-8. [Google Scholar]
- Adkesson, M.J.; Zdziarski, J.M.; Little, S.E. Atoxoplasmosis in tanagers. J. Zoo Wildl. Med. 2005, 36, 265–272. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.R.; de Souza, T.D.; Mol, J.P.S.; Flecher, M.C.; Hiura, E.; Santos, R.L. Pathological and molecular characterization of systemic isosporosis (atoxoplasmosis) in captive green-winged saltator (Saltator similis). Vet. Parasitol. 2018, 255, 98–101. [Google Scholar] [CrossRef]
- Maslin, W.R.; Latimer, K.S. Atoxoplasmosis in canary fledglings: Severe lymphocytic enteritis with preferential parasitism of B lymphocytes. Avian Dis. 2009, 53, 473–476. [Google Scholar] [CrossRef]
- Sánchez-Cordón, P.J.; Gómez-Villamandos, J.C.; Gutiérrez, J.; Sierra, M.A.; Pedrera, M.; Bautista, M.J. Atoxoplasma spp. infection in captive canaries (Serinus canaria). J. Vet. Med. Ser. A 2007, 54, 23–26. [Google Scholar] [CrossRef]
- Trupkiewicz, J.; Garner, M.M.; Juan-Sallés, C. Passeriformes, Caprimulgiformes, Coraciiformes, Piciformes, Bucerotiformes, and Apodiformes. In Pathology of Wildlife and Zoo Animals; Elsevier: Amsterdam, The Netherlands, 2018; pp. 799–823. ISBN 978-0-12-805306-5. [Google Scholar]
- Kubiski, S.V.; Witte, C.; Burchell, J.A.; Conradson, D.; Zmuda, A.; Barbon, A.R.; Vilches-Moure, J.G.; Felt, S.A.; Rideout, B.A. Mitochondrial gene diversity and host specificity of Isospora in passerine birds. Front. Vet. Sci. 2022, 9, 847030. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Brice, B.; Elliot, A.; Ryan, U.; Yang, R. Isospora coronoideae n. sp. (Apicomplexa: Eimeriidae) from the Australian raven (Corvus coronoides) (Passeriformes: Corvidae) (Linnaeus, 1758) in Western Australia. Parasitol. Res. 2019, 118, 2399–2408. [Google Scholar] [CrossRef] [PubMed]
- Rejman, E.E.; Hák-Kovács, K.; Barta, J.R. The first Isospora species (Apicomplexa: Eimeriidae) described from the northern yellow-shafted flicker (Colaptes auratus luteus) in Ontario, Canada. Acta Parasitol. 2022, 67, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Trefancová, A.; Kvičerová, J. Isospora svecica sp. n. (Apicomplexa: Eimeriidae), a new species of coccidium from the white-spotted bluethroat Luscinia svecica cyanecula (Aves: Passeriformes: Muscicapidae). Parasitol. Res. 2019, 118, 3043–3051. [Google Scholar] [CrossRef] [PubMed]
- Labbé, A. Sporozoa. In Das Tierreich. Eine Zusammenstellung und Kennzeichnung der Rezenten Tierformen; Schulze, F.E., Butschli, O., Eds.; Walter de Gruyter: Berlin, Germany, 1899; p. 180. [Google Scholar]
- Barta, J.R.; Martin, D.S.; Carreno, R.A.; Siddall, M.E.; Profous-Juchelka, H.; Hozza, M.; Ann Powles, M.; Sundermann, C. Molecular phylogeny of the other tissue Coccidia: Lankesterella and Caryospora. J. Parasitol. 2001, 87, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Desser, S.S.; Siddall, M.E.; Barta, J.R. Ultrastructural observations on the developmental stages of Lankesterella minima (Apicomplexa) in experimentally infected Rana catesbeiana tadpoles. J. Parasitol. 1990, 76, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Gericota, B.; Garner, M.M.; Barr, B.; Nordhausen, R.; Larsen, R.S.; Lowenstine, L.J.; Murphy, B.G. Morphologic, immunohistochemical, and molecular characterization of a novel Lankesterella protozoan in two white’s tree frogs (Litoria caerulea). J. Zoo Wildl. Med. 2010, 41, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Lainson, R.; Paperna, I. Light and electron microscope study of a Lankesterella petiti n. sp., (Apicomplexa: Lankesterellidae) infecting Bufo marinus (Amphibia: Anura) in Pará, North Brazil. Parasite 1995, 2, 307–313. [Google Scholar] [CrossRef]
- Paperna, I.; Bastien, P.; Chavatte, J.-M.; Landau, I. Lankesterella poeppigii n. sp. (Apicomplexa, Lankesterellidae) de Bufo poeppigii (Tschudi, 1845) del Perú. Rev. Peru. Biol. 2009, 16, 165–168. [Google Scholar] [CrossRef]
- Paperna, I.; Martin, C. The Development and fine structure of Lankesterella cf. dicroglossi (Apicomplexa: Lankesterellidae) infecting frogs in Niger, West Africa. Folia Parasitol. 2001, 48, 178–186. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Lin, T.-S.; Huang, W.-W.; Lee, H.-Y.; Shih, C.-H.; Wu, Y.-C.; Huang, C.-C.; Chen, T.-H. Reevaluation of hemoparasites in the black spiny-tailed iguana (Ctenosaura similis) with the first pathological and molecular characterizations of Lankesterella desseri n. sp. and redescription of Hepatozoon gamezi. Microorganisms 2023, 11, 2374. [Google Scholar] [CrossRef] [PubMed]
- Hajiyan, R.; Javanbakht, H. Molecular characterization of haemoparasites, genus Lankestrella (Apicomplexa: Coccidia) in two lizard species from Iran. J. Wildl. Biodivers. 2024, 8, 192–202. [Google Scholar] [CrossRef]
- Megía-Palma, R.; Martínez, J.; Paranjpe, D.; D’Amico, V.; Aguilar, R.; Palacios, M.G.; Cooper, R.; Ferri-Yáñez, F.; Sinervo, B.; Merino, S. Phylogenetic analyses reveal that Schellackia parasites (Apicomplexa) detected in American lizards are closely related to the genus Lankesterella: Is the range of Schellackia restricted to the Old World? Parasites Vectors 2017, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Megía-Palma, R.; Martínez, J.; Cuervo, J.J.; Belliure, J.; Jiménez-Robles, O.; Gomes, V.; Cabido, C.; Pausas, J.G.; Fitze, P.S.; Martín, J.; et al. Molecular evidence for host–parasite co-speciation between lizards and Schellackia Parasites. Int. J. Parasitol. 2018, 48, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Telford, S.R. The Hemoparasites of the Reptilia; CRC: Boca Raton, FL, USA, 2009. [Google Scholar]
- Saravana Bhavan Venkatachalam, A.K.; Čepička, I.; Hrazdilová, K.; Svobodová, M. Host specificity of passerine Lankesterella (Apicomplexa: Coccidia). Eur. J. Protistol. 2023, 90, 126007. [Google Scholar] [CrossRef] [PubMed]
- Šujanová, A.; Špitalská, E.; Václav, R. Seasonal dynamics and diversity of haemosporidians in a natural woodland bird community in Slovakia. Diversity 2021, 13, 439. [Google Scholar] [CrossRef]
- Chvala, S.; Bakonyi, T.; Bukovsky, C.; Meister, T.; Brugger, K.; Rubel, F.; Nowotny, N.; Weissenböck, H. Monitoring of Usutu virus activity and spread by using dead bird surveillance in Austria, 2003–2005. Vet. Microbiol. 2007, 122, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Himmel, T.; Harl, J.; Matt, J.; Weissenböck, H. A Citizen science-based survey of avian mortality focusing on haemosporidian infections in wild passerine birds. Malar. J. 2021, 20, 417. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G.; Iezhova, T.A.; Križanauskienė, A.; Palinauskas, V.; Sehgal, R.N.M.; Bensch, S. A Comparative analysis of microscopy and PCR-based detection methods for blood parasites. J. Parasitol. 2008, 94, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Weissenböck, H.; Kolodziejek, J.; Url, A.; Lussy, H.; Rebel-Bauder, B.; Nowotny, N. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, Central Europe. Emerg. Infect. Dis. 2002, 8, 652–655. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Bensch, S.; Hellgren, O.; Pérez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z. DAMBE: Data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Dinhopl, N.; Mostegl, M.M.; Richter, B.; Nedorost, N.; Maderner, A.; Fragner, K.; Weissenböck, H. In situ hybridisation for the detection of Leishmania species in paraffin wax-embedded canine tissues using a digoxigenin-labelled oligonucleotide probe. Vet. Rec. 2011, 169, 525. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.; Paperna, I. Proliferative visceral Isospora (atoxoplasmosis) with morbid impact on the Israeli sparrow Passer domesticus biblicus Hartert, 1904. Parasitol Res. 2008, 103, 493–499. [Google Scholar] [CrossRef]
- Ogedengbe, M.E.; Brash, M.; Barta, J.R. The complete mitochondrial genome sequence of an Isospora sp. (Eimeriidae, Eucoccidiorida, Coccidiasina, Apicomplexa) causing systemic coccidiosis in domestic canaries (Serinus canaria Linn.). Mitochondrial DNA Part A 2016, 27, 3315–3317. [Google Scholar] [CrossRef]
- Mohr, F.; Betson, M.; Quintard, B. Investigation of the presence of Atoxoplasma spp. in blue-crowned laughingthrush (Dryonastes courtoisi) adults and neonates. J. Zoo Wildl. Med. 2017, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Partington, C.J.; Gardiner, C.H.; Fritz, D.; Phillips, L.G.; Montali, R.J. Atoxoplasmosis in Bali mynahs (Leucopsar rothschildi). J. Zoo Wildl. Med. 1989, 20, 328–335. [Google Scholar]
- Illera, J.C.; Fernández-Álvarez, Á.; Hernández-Flores, C.N.; Foronda, P. Unforeseen biogeographical patterns in a multiple parasite system in Macaronesia. J. Biogeogr. 2015, 42, 1858–1870. [Google Scholar] [CrossRef]
- Svobodová, M. Isospora, Caryospora and Eimeria (Apicomplexa: Passeriform birds from Czech Republic. Acta Protozool. 1994, 33, 101–108. [Google Scholar]
- Trefancová, A.; Mácová, A.; Kvičerová, J. Isosporan oocysts in the faeces of bank voles (Myodes glareolus; Arvicolinae, Rodentia): Real parasites, or pseudoparasites? Protist 2019, 170, 104–120. [Google Scholar] [CrossRef] [PubMed]
- Megía-Palma, R.; Martínez, J.; Nasri, I.; Cuervo, J.J.; Martín, J.; Acevedo, I.; Belliure, J.; Ortega, J.; García-Roa, R.; Selmi, S.; et al. Phylogenetic relationships of Isospora, Lankesterella, and Caryospora species (Apicomplexa: Eimeriidae) infecting lizards. Org. Divers. Evol. 2016, 16, 275–288. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Dubey, J.P.; Santín-Durán, M. Coccidia and other protozoa. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 1015–1027. ISBN 978-1-119-35085-9. [Google Scholar]
- Barbón, A.R.; López, J.; Jamriška, J.; Thomasson, A.; Braun, J.; Stidworthy, M.F. Clinical and pathological aspects of systemic Isospora infection in blue-crowned laughing thrushes (Garrulax courtoisi) at Jersey Zoo. J. Avian Med. Surg. 2019, 33, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Cushing, T.L.; Schat, K.A.; States, S.L.; Grodio, J.L.; O’Connell, P.H.; Buckles, E.L. Characterization of the host response in systemic isosporosis (atoxoplasmosis) in a colony of captive American goldfinches (Spinus tristis) and house sparrows (Passer domesticus). Vet. Pathol. 2011, 48, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Laird, M. Atoxoplasma paddae (Aragão) from several South Pacific silvereyes (Zosteropidae) and a New Zealand rail. J. Parasitol. 1959, 45, 47–52. [Google Scholar] [CrossRef]
- Middleton, A.L.A.; Julian, R.J. Lymphoproliferative disease in the american goldfinch, Carduelis tristis. J. Wildl. Dis. 1983, 19, 280–285. [Google Scholar] [CrossRef]
- Quiroga, M.I.; Aleman, N.; Vazquez, S.; Nieto, J.M. Diagnosis of atoxoplasmosis in a canary (Serinus canarius) by histopathologic and ultrastructural examination. Avian Dis. 2000, 44, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, P.L.; Bowerman, W.J. Hemoparasites in Oregon spotted frogs (Rana pretiosa) from Central Oregon, USA. J. Wildl. Dis. 2008, 44, 464–468. [Google Scholar] [CrossRef] [PubMed]






| Host Family | Host Species | N | IR (%) | Isospora Lineage | IR (%) | Lankesterella Lineage |
|---|---|---|---|---|---|---|
| Acrocephalidae | Acrocephalus arundinaceus | 3 | - | - | ||
| Acrocephalus melanopogon | 9 | - | 11.1 | kACRSCH01 (1) | ||
| Acrocephalus palustris | 19 | 5.3 | iACRPAL01 (1) | 10.5 | kACRPAL02 (2) | |
| Acrocephalus schoenobaenus | 8 | - | 37.5 | kACRSCH01 (2), kACRSCH02 (2) | ||
| Acrocephalus scirpaceus | 22 | 9.1 | iACRSCI01 (1), iACRSCI02 (1), iACRPAL01 (1) | - | ||
| Acrocephalus sp. | 1 | - | - | |||
| Hippolais icterina | 1 | - | - | |||
| Aegithalidae | Aegithalos caudatus | 1 | - | - | ||
| Alaudidae | Calandrella brachydactyla | 1 | - | - | ||
| Certhiidae | Certhia brachydactyla | 1 | - | - | ||
| Corvidae | Garrulus glandarius | 5 | - | - | ||
| Emberizidae | Emberiza citrinella | 10 | 20.0 | iEMBCIR01 (1), iEMBCIR02 (1) | - | |
| Emberiza schoeniclus | 7 | - | - | |||
| Fringillidae | Carduelis carduelisF | 3 | - | - | ||
| Coccothraustes coccothraustes | 19 | - | - | |||
| Fringilla coelebs | 6 | 16.6 | iFRICOE01 (1) | - | ||
| Pyrrhula pyrrhula | 7 | - | - | |||
| Hirundinidae | Delichon urbicum | 3 | - | 33.3 | kHIRRUS01 (1) | |
| Hirundinidae sp. | 1 | - | - | |||
| Hirundo rustica | 43 | - | 14.0 | kHIRRUS01 (4), kSYLATR01 (2) | ||
| Hirundo sp. | 1 | - | - | |||
| Laniidae | Lanius collurio | 7 | - | 42.9 | kLANCOL01 (2), kLANCOL02 (1) | |
| Locustellidae | Locustella luscinioides | 9 | - | - | ||
| Locustella naevia | 1 | - | - | |||
| Motacillidae | Motacilla alba | 4 | - | 50.0 | kMOTALB01 (2) | |
| Motacilla flava | 1 | - | - | |||
| Muscicapidae | Erithacus rubecula | 104 | 5.8 | iERIRUB01 (1), iERIRUB02 (3), iERIRUB03 (1), iERIRUB04 (1) | 21.2 | kERIRUB01 (21), kERIRUB02 (1), kSYLATR01 (1) |
| Ficedula hypoleuca | 11 | - | 45.5 | kFICHYP01 (4), kFICHYP02 (1) | ||
| Luscinia luscinia | 1 | 100.0 | iLUSLUS01 (1) | - | ||
| Luscinia megarynchos | 5 | - | - | |||
| Muscicapa striata | 1 | - | - | |||
| Phoenicurus ochruros | 8 | 62.5 | iPHOOCH01 (4), iPHOOCH02 (1) | - | ||
| Phoenicurus phoenicurus | 3 | - | - | |||
| Phoenicurus sp. | 1 | - | - | |||
| Saxicola rubicola | 1 | - | - | |||
| Panuridae | Panurus biarmicus | 4 | - | - | ||
| Paridae | Cyanistes caeruleus | 65 | - | 1.5 | kCYACAE01 (1) | |
| Parus major | 86 | 3.5 | iPARMAJ01 (1), iPASDOM01 (1), iPASDOM02 (1) | 7.0 | kPARMAJ01 (4), kPARMAJ02 (2) | |
| Parus sp. | 1 | - | - | |||
| Periparus ater | 1 | - | - | |||
| Poecile palustris | 8 | - | - | |||
| Passeridae | Passer domesticus | 46 | 50.0 | iPASDOM01 (5), iPASDOM02 (7), iPASDOM03 (8), iPASDOM04 (1), iPASDOM05 (1), iPASDOM06 (1) | 6.5 | kPASDOM01 (1), kPASDOM03 (1), kPASDOM04 (1) |
| Passer montanus | 8 | 12.5 | iPASMON01 (1) | - | ||
| Passer sp. | 3 | 100.0 | iPASDOM01 (1), iPASDOM02 (2) | 33.3 | kPASDOM02 (1) | |
| Phylloscopidae | Phylloscopus collybita | 18 | 11.1 | iPHYCOL01 (2) | 5.6 | kPHYCOL01 (1) |
| Phylloscopus sibilatrix | 4 | - | - | |||
| Phylloscopus trochiloides | 2 | - | - | |||
| Phylloscopus trochilus | 18 | 16.7 | iPHYCOL01 (2), iPHYTRO01 (1) | 5.6 | kPHYCOL02 (1) | |
| Prunellidae | Prunella modularis | 14 | - | 21.4 | kPRUMOD01 (3) | |
| Regulidae | Regulus ignicapilla | 3 | - | |||
| Regulus regulus | 11 | 27.3 | iREGREG01 (2), iREGREG02 (1) | 9.1 | kREGREG01 (1) | |
| Sylviidae | Sylvia atricapilla | 118 | 2.5 | iSYLATR01 (1), iSYLATR02 (2) | 17.8 | kSYLATR01 (18), kSYLATR02 (1), kACRSCH01 (1), kPARMAJ01 (1) |
| Sylvia borin | 7 | - | - | |||
| Curruca communis | 9 | - | 11.1 | kSYLCOM01 (1) | ||
| Curruca curruca | 10 | - | 20.0 | kSYLCUR01 (2) | ||
| Sylvia sp. | 1 | - | - | |||
| Troglodytidae | Troglodytes troglodytes | 5 | - | - | ||
| Turdidae | Turdus merula | 28 | - | 7.1 | kTURMER01 (1), kSYLATR01 (1) | |
| Turdus philomelos | 12 | - | - | |||
| Captive birds | ||||||
| Cardinalidae | Passerina ciris | 1 | 100.0 | iPASCIR01 (1) | - | |
| Estrildidae | Padda oryzivora | 1 | 100.0 | iPADORY01 (1) | - | |
| Fringillidae | Carduelis carduelis | 1 | 100.0 | iSYLATR01 (1) | - | |
| Spinus cucullatus | 3 | - | - | |||
| Serinus canaria | 3 | 33.3 | iSERCAN01 (1) | - | ||
| Spinus xanthogastrus | 3 | - | - | |||
| Spinus yarrellii | 3 | 33.3 | iSPIYAR01 (1) | - |
| Host Family | Host Species | Host-ID | CISH | HE | LU | LI | SP | KI | BR | MU | GI | IN | TR | ES | TE | OV | BU | PC | CR | BM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acrocephalidae | Acrocephalus palustris | AH1899 | pos | + | n/a | + | n/a | + | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Acrocephalus scirpaceus | AH1896 | pos | + | + | + | + | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Cardinalidae | Passerina ciris (c) | B1276/12 | pos | + | ++ | +++ | +++ | ++ | + | + | ++ | + | n/a | n/a | n/a | ++ | n/a | n/a | ++ | +++ |
| Estrildidae | Padda oryzivora (c) | B727/17 | pos | + | + | +++ | +++ | + | + | + | ++ | +++ | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a |
| Fringillidae | Carduelis carduelis | B421/13 | pos | n/a | n/a | +++ | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Serinus canaria (c) | B1277/12 | neg | n/a | n/a | n/a | n/a | n/a | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Spinus yarrellii (c) | T331/12 | pos | − | − | ++ | +++ | + | − | n/a | + | + | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | |
| Muscicapidae | Erithacus rubecula | AH1980 | pos | − | − | + | + | − | − | − | − | +++ | n/a | n/a | n/a | − | n/a | − | n/a | n/a |
| Erithacus rubecula | AH2000 | neg | − | − | − | n/a | − | − | − | − | − | n/a | n/a | − | n/a | n/a | n/a | n/a | n/a | |
| Phoenicurus ochruros | AH0792 | pos | − | − | + | + | n/a | − | n/a | − | − | − | − | n/a | n/a | n/a | − | n/a | n/a | |
| Phoenicurus ochruros | AH1977 | pos | − | n/a | + | n/a | − | − | − | − | + | n/a | n/a | n/a | n/a | n/a | − | n/a | n/a | |
| Phoenicurus ochruros | AH1988 | pos | − | − | − | n/a | + | − | − | − | + | n/a | − | n/a | n/a | n/a | n/a | n/a | n/a | |
| Phoenicurus ochruros | AH2009 | pos | − | − | + | n/a | − | − | − | − | + | − | − | − | n/a | n/a | n/a | n/a | n/a | |
| Paridae | Parus major | AH1998 | neg | − | − | − | − | − | − | − | − | − | n/a | n/a | n/a | n/a | − | n/a | n/a | n/a |
| Parus major | AH1999 | pos | − | − | + | n/a | − | − | − | − | ++ | n/a | n/a | n/a | − | n/a | n/a | n/a | n/a | |
| Passeridae | Passer domesticus | AH0597 | pos | − | − | n/a | +++ | − | − | n/a | ++ | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Passer domesticus | AH1553 | neg | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | − | n/a | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH1557 | pos | − | n/a | − | + | n/a | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH1559 | pos | + | − | + | + | + | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH1562 | pos | − | + | + | +++ | ++ | − | n/a | ++ | n/a | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH1566 | pos | − | + | ++ | +++ | + | − | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH1570 | pos | − | n/a | ++ | n/a | + | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passer domesticus | AH2031 | pos | + | − | − | n/a | − | − | + | + | + | − | − | − | n/a | n/a | n/a | n/a | n/a | |
| Passer montanus | AH0761 | pos | − | + | + | + | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passer sp. | AH1989 | pos | − | + | − | + | − | − | + | + | + | n/a | − | n/a | n/a | − | + | n/a | n/a | |
| Passer sp. | AH2026 | pos | + | + | + | n/a | − | − | + | − | + | − | − | − | n/a | n/a | n/a | n/a | n/a |
| Host Family | Host Species | Host-ID | CISH | HE | LU | LI | SP | KI | BR | MU | GI | IN | TR | ES | TE | OV | BU | PC | CR | BM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acrocephalidae | Acrocephalus melanopogon | AH1591 | pos | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Emberizidae | Emberiza citrinella | AH1888 | neg | − | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Hirundinidae | Hirundo rustica | AH2003 | pos | − | − | − | n/a | − | + | − | − | n/a | − | − | n/a | n/a | − | n/a | n/a | n/a |
| Motacillidae | Motacilla alba | AH1897 | pos | + | + | + | + | + | + | + | n/a | n/a | n/a | n/a | + | n/a | n/a | − | n/a | n/a |
| Motacilla alba | AH1969 | pos | + | n/a | + | + | + | + | − | + | + | n/a | n/a | + | n/a | n/a | − | n/a | n/a | |
| Muscicapidae | Erithacus rubecula | AH1580 | neg | n/a | n/a | n/a | n/a | n/a | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Erithacus rubecula | AH1634 | pos | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Erithacus rubecula | AH1966 | pos | − | + | + | n/a | − | − | − | − | − | n/a | − | n/a | − | n/a | − | n/a | n/a | |
| Erithacus rubecula | AH1970 | neg | − | n/a | − | n/a | − | − | − | − | − | n/a | − | n/a | n/a | n/a | − | n/a | n/a | |
| Erithacus rubecula | AH2139 | neg | − | − | − | n/a | − | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | |
| Erithacus rubecula | AH2148 | pos | − | n/a | n/a | n/a | n/a | n/a | + | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Paridae | Cyanistes caeruleus | AH2043 | neg | n/a | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Parus major | AH2039 | neg | − | − | − | n/a | − | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | |
| Passeridae | Passer domesticus | AH1569 | neg | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Passer domesticus | AH0462 | pos | − | − | − | n/a | − | − | n/a | + | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| Regulidae | Regulus regulus | AH2146 | neg | − | − | − | n/a | − | − | − | − | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a |
| Sylviidae | Sylvia atricapilla | AH1894 | pos | − | − | − | + | − | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Sylvia atricapilla | AH1905 | pos | − | + | − | − | − | − | − | n/a | n/a | n/a | n/a | − | n/a | n/a | n/a | n/a | n/a | |
| Sylvia atricapilla | AH1991 | pos | + | + | − | + | + | − | − | − | n/a | n/a | n/a | − | n/a | − | n/a | n/a | n/a | |
| Curruca communis | AH1900 | pos | − | + | − | − | − | + | − | n/a | n/a | n/a | n/a | − | n/a | n/a | n/a | n/a | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keckeisen, C.; Šujanová, A.; Himmel, T.; Matt, J.; Nedorost, N.; Chagas, C.R.F.; Weissenböck, H.; Harl, J. Isospora and Lankesterella Parasites (Eimeriidae, Apicomplexa) of Passeriform Birds in Europe: Infection Rates, Phylogeny, and Pathogenicity. Pathogens 2024, 13, 337. https://doi.org/10.3390/pathogens13040337
Keckeisen C, Šujanová A, Himmel T, Matt J, Nedorost N, Chagas CRF, Weissenböck H, Harl J. Isospora and Lankesterella Parasites (Eimeriidae, Apicomplexa) of Passeriform Birds in Europe: Infection Rates, Phylogeny, and Pathogenicity. Pathogens. 2024; 13(4):337. https://doi.org/10.3390/pathogens13040337
Chicago/Turabian StyleKeckeisen, Carina, Alžbeta Šujanová, Tanja Himmel, Julia Matt, Nora Nedorost, Carolina R. F. Chagas, Herbert Weissenböck, and Josef Harl. 2024. "Isospora and Lankesterella Parasites (Eimeriidae, Apicomplexa) of Passeriform Birds in Europe: Infection Rates, Phylogeny, and Pathogenicity" Pathogens 13, no. 4: 337. https://doi.org/10.3390/pathogens13040337
APA StyleKeckeisen, C., Šujanová, A., Himmel, T., Matt, J., Nedorost, N., Chagas, C. R. F., Weissenböck, H., & Harl, J. (2024). Isospora and Lankesterella Parasites (Eimeriidae, Apicomplexa) of Passeriform Birds in Europe: Infection Rates, Phylogeny, and Pathogenicity. Pathogens, 13(4), 337. https://doi.org/10.3390/pathogens13040337