Conjugation Operons in Gram-Positive Bacteria with and without Antitermination Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Media and Oligonucleotides
2.2. Construction of Plasmids and Strains
2.3. Transformation
2.4. Flow Cytometry
2.5. Identification of Putative Termination Sequences
3. Results
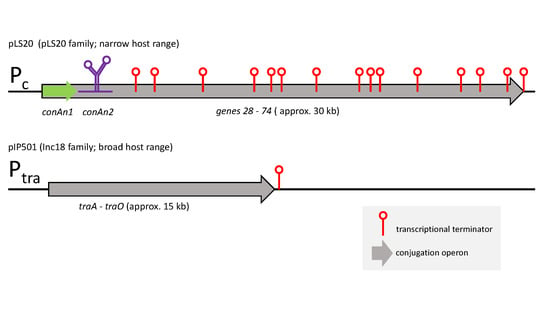
3.1. Highly Similar Conjugation Operons of Inc18 Plasmids pIP501, pRE25 and pAMβ1 Contain Two Putative Transcriptional Terminators
3.2. Putative Terminator Ter1 Present in Conjugation Operons of pIP501, pAMβ1 and pRE25 Is Not Functional In Vivo
3.3. Additional Evidence That pIP501, pAMβ1 and pRE25 Do Not Contain a Processive Antitermination System: Their Conjugation Operons Start with Relaxase Gene
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabezon, E.; Ripoll-Rozada, J.; Pena, A.; De la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2014, 39, 81–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goessweiner-Mohr, N.; Arends, K.; Keller, W.; Grohmann, E. Conjugation in Gram-Positive Bacteria. Microbiol. Spectr. 2014, 2, PLAS-0004-2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazel, D.; Davies, J. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 1999, 56, 742–754. [Google Scholar] [CrossRef]
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; De la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [Green Version]
- Bravo, A.; Ruiz-Cruz, S.; Alkorta, I.; Espinosa, M. When Humans Met Superbugs: Strategies to Tackle Bacterial Resistances to Antibiotics. Biomol. Concepts 2018, 9, 216–226. [Google Scholar] [CrossRef]
- Breuer, R.J.; Hirt, H.; Dunny, G.M. Mechanistic Features of the Enterococcal pCF10 Sex Pheromone Response and the Biology of Enterococcus faecalis in Its Natural Habitat. J. Bacteriol. 2018, 200, e00733-17. [Google Scholar] [CrossRef] [Green Version]
- Dunny, G.M. Enterococcal sex pheromones: Signaling, social behavior, and evolution. Annu. Rev. Genet. 2013, 47, 457–482. [Google Scholar] [CrossRef]
- Kohler, V.; Keller, W.; Grohmann, E. Regulation of Gram-Positive Conjugation. Front. Microbiol. 2019, 10, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grohmann, E.; Goessweiner-Mohr, N.; Brantl, S. DNA-Binding Proteins Regulating pIP501 Transfer and Replication. Front. Mol. Biosci. 2016, 3, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisniewski, J.A.; Rood, J.I. The Tcp conjugation system of Clostridium perfringens. Plasmid 2017, 91, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.; Han, X.; Lyras, D.; Rood, J.I. Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens. Plasmid 2018, 99, 32–39. [Google Scholar] [CrossRef]
- Singh, P.K.; Meijer, W.J. Diverse regulatory circuits for transfer of conjugative elements. FEMS Microbiol. Lett. 2014, 358, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Meijer, W.J.J.; Boer, D.R.; Ares, S.; Alfonso, C.; Rojo, F.; Luque-Ortega, J.R.; Wu, L.J. Multiple Layered Control of the Conjugation Process of the Bacillus subtilis Plasmid pLS20. Front. Mol. Biosci. 2021, 8, 648468. [Google Scholar] [CrossRef]
- Dunny, G.M.; Berntsson, R.P. Enterococcal Sex Pheromones: Evolutionary Pathways to Complex, Two-Signal Systems. J. Bacteriol. 2016, 198, 1556–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lybecker, M.; Bilusic, I.; Raghavan, R. Pervasive transcription: Detecting functional RNAs in bacteria. Transcription 2014, 5, e944039. [Google Scholar] [CrossRef] [Green Version]
- Wade, J.T.; Grainger, D.C. Spurious transcription and its impact on cell function. Transcription 2018, 9, 182–189. [Google Scholar] [CrossRef]
- Wade, J.T.; Grainger, D.C. Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014, 12, 647–653. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W.R. Widespread Antisense Transcription in Prokaryotes. Microbiol. Spectr. 2018, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Vangeloff, A.D.; Landick, R. Bacterial transcription terminators: The RNA 3′-end chronicles. J. Mol. Biol. 2011, 412, 793–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Washburn, R.S.; Gottesman, M.E. Regulation of transcription elongation and termination. Biomolecules 2015, 5, 1063–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porrua, O.; Libri, D. Transcription termination and the control of the transcriptome: Why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015, 16, 190–202. [Google Scholar] [CrossRef]
- Porrua, O.; Boudvillain, M.; Libri, D. Transcription Termination: Variations on Common Themes. Trends Genet. 2016, 32, 508–522. [Google Scholar] [CrossRef]
- Ray-Soni, A.; Bellecourt, M.J.; Landick, R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu. Rev. Biochem. 2016, 85, 319–347. [Google Scholar] [CrossRef] [Green Version]
- Mitra, P.; Ghosh, G.; Hafeezunnisa, M.; Sen, R. Rho Protein: Roles and Mechanisms. Annu. Rev. Microbiol. 2017, 71, 687–709. [Google Scholar] [CrossRef]
- Greenblatt, J.; McLimont, M.; Hanly, S. Termination of transcription by nusA gene protein of Escherichia coli. Nature 1981, 292, 215–220. [Google Scholar] [CrossRef]
- Mondal, S.; Yakhnin, A.V.; Sebastian, A.; Albert, I.; Babitzke, P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 2016, 1, 15007. [Google Scholar] [CrossRef]
- Miguel-Arribas, A.; Val-Calvo, J.; Gago-Cordoba, C.; Izquierdo, J.M.; Abia, D.; Wu, L.J.; Errington, J.; Meijer, W.J.J. A novel bipartite antitermination system widespread in conjugative elements of Gram-positive bacteria. Nucleic Acids Res. 2021, 49, 5553–5567. [Google Scholar] [CrossRef]
- Val-Calvo, J.; Miguel-Arribas, A.; Abia, D.; Wu, L.J.; Meijer, W.J.J. pLS20 is the archetype of a new family of conjugative plasmids harboured by Bacillus species. NAR Genom. Bioinform. 2021, 3, lqab096. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, R.A.; Gottesman, M.E. Processive antitermination. J. Bacteriol. 1999, 181, 359–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodson, J.R.; Winkler, W.C. Processive Antitermination. Microbiol. Spectr. 2018, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- King, R.A.; Banik-Maiti, S.; Jin, D.J.; Weisberg, R.A. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell 1996, 87, 893–903. [Google Scholar] [CrossRef] [Green Version]
- Sen, R.; King, R.A.; Weisberg, R.A. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol. Cell 2001, 7, 993–1001. [Google Scholar] [CrossRef]
- Wang, B.; Artsimovitch, I. NusG, an Ancient Yet Rapidly Evolving Transcription Factor. Front. Microbiol. 2020, 11, 619618. [Google Scholar] [CrossRef]
- Komissarova, N.; Velikodvorskaya, T.; Sen, R.; King, R.A.; Banik-Maiti, S.; Weisberg, R.A. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol. Cell 2008, 31, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Irnov, I.; Winkler, W.C. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol. Microbiol. 2010, 76, 559–575. [Google Scholar] [CrossRef]
- Banik-Maiti, S.; King, R.A.; Weisberg, R.A. The antiterminator RNA of phage HK022. J. Mol. Biol. 1997, 272, 677–687. [Google Scholar] [CrossRef]
- Gago-Cordoba, C.; Val-Calvo, J.; Miguel-Arribas, A.; Serrano, E.; Singh, P.K.; Abia, D.; Wu, L.J.; Meijer, W.J.J. Surface Exclusion Revisited: Function Related to Differential Expression of the Surface Exclusion System of Bacillus subtilis Plasmid pLS20. Front. Microbiol. 2019, 10, 1502. [Google Scholar] [CrossRef]
- Mikalsen, T.; Pedersen, T.; Willems, R.; Coque, T.M.; Werner, G.; Sadowy, E.; van Schaik, W.; Jensen, L.B.; Sundsfjord, A.; Hegstad, K. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 2015, 16, 282. [Google Scholar] [CrossRef] [Green Version]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, V.; Vaishampayan, A.; Grohmann, E. Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms. Plasmid 2018, 99, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Volante, A.; Soberon, N.E.; Ayora, S.; Alonso, J.C. The Interplay between Different Stability Systems Contributes to Faithful Segregation: Streptococcus pyogenes pSM19035 as a Model. Microbiol. Spectr. 2014, 2, PLAS-0007-2013. [Google Scholar] [CrossRef] [Green Version]
- Icgen, B. VanA-Type MRSA (VRSA) Emerged in Surface Waters. Bull. Environ. Contam. Toxicol. 2016, 97, 359–366. [Google Scholar] [CrossRef]
- Zhu, W.; Clark, N.; Patel, J.B. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob. Agents Chemother. 2013, 57, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, V.S.; Zervos, M.J.; Kaye, K.S.; Tosh, P.K.; Arshad, S.; Hayakawa, K.; Kallen, A.J.; McDougal, L.K.; Limbago, B.M.; Guh, A.Y. Prevalence of and risk factors for vancomycin-resistant Staphylococcus aureus precursor organisms in Southeastern Michigan. Infect. Control. Hosp. Epidemiol. 2014, 35, 1531–1534. [Google Scholar] [CrossRef]
- Jain, A.; Srivastava, P. Broad host range plasmids. FEMS Microbiol. Lett. 2013, 348, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Rosvoll, T.C.; Pedersen, T.; Sletvold, H.; Johnsen, P.J.; Sollid, J.E.; Simonsen, G.S.; Jensen, L.B.; Nielsen, K.M.; Sundsfjord, A. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 2010, 58, 254–268. [Google Scholar] [CrossRef] [Green Version]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Singh, P.K.; Ramachandran, G.; Ramos-Ruiz, R.; Peiro-Pastor, R.; Abia, D.; Wu, L.J.; Meijer, W.J. Mobility of the Native Bacillus subtilis Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling. PLoS Genet. 2013, 9, e1003892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautheret, D.; Lambert, A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001, 313, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Macke, T.J.; Ecker, D.J.; Gutell, R.R.; Gautheret, D.; Case, D.A.; Sampath, R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001, 29, 4724–4735. [Google Scholar] [CrossRef] [PubMed]
- Kingsford, C.L.; Ayanbule, K.; Salzberg, S.L. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007, 8, R22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurenbach, B.; Kopec, J.; Magdefrau, M.; Andreas, K.; Keller, W.; Bohn, C.; Abajy, M.Y.; Grohmann, E. The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology 2006, 152, 637–645. [Google Scholar] [CrossRef] [Green Version]
- Komissarova, N.; Becker, J.; Solter, S.; Kireeva, M.; Kashlev, M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 2002, 10, 1151–1162. [Google Scholar] [CrossRef]
- D’Aubenton Carafa, Y.; Brody, E.; Thermes, C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990, 216, 835–858. [Google Scholar] [CrossRef]
- Kurenbach, B.; Grothe, D.; Farias, M.E.; Szewzyk, U.; Grohmann, E. The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase. J. Bacteriol. 2002, 184, 1801–1805. [Google Scholar] [CrossRef] [Green Version]
- Barth, P.T.; Grinter, N.J.; Bradley, D.E. Conjugal transfer system of plasmid RP4: Analysis by transposon 7 insertion. J. Bacteriol. 1978, 133, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Ramachandran, G.; Duran-Alcalde, L.; Alonso, C.; Wu, L.J.; Meijer, W.J. Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 2012, 14, 2812–2825. [Google Scholar] [CrossRef]
- Almeida, L.M.; Gaca, A.; Bispo, P.M.; Lebreton, F.; Saavedra, J.T.; Silva, R.A.; Basilio-Junior, I.D.; Zorzi, F.M.; Filsner, P.H.; Moreno, A.M.; et al. Coexistence of the Oxazolidinone Resistance-Associated Genes cfr and optrA in Enterococcus faecalis From a Healthy Piglet in Brazil. Front Public Health 2020, 8, 518. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S.; Behnke, D.; Alonso, J.C. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAMβ1 and pSM19035. Nucleic Acids Res. 1990, 18, 4783–4790. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Murray, P.R.; Huskins, W.C.; Jernigan, J.A.; McDonald, L.C.; Clark, N.C.; Anderson, K.F.; McDougal, L.K.; Hageman, J.C.; Olsen-Rasmussen, M.; et al. Dissemination of an Enterococcus Inc18-Like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 4314–4320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurenbach, B.; Bohn, C.; Prabhu, J.; Abudukerim, M.; Szewzyk, U.; Grohmann, E. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 2003, 50, 86–93. [Google Scholar] [CrossRef]
- Flannagan, S.E.; Chow, J.W.; Donabedian, S.M.; Brown, W.J.; Perri, M.B.; Zervos, M.J.; Ozawa, Y.; Clewell, D.B. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob. Agents Chemother. 2003, 47, 3954–3959. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Wan, K.; Wang, H.H. High-frequency conjugation system facilitates biofilm formation and pAMbeta1 transmission by Lactococcus lactis. Appl. Environ. Microbiol. 2005, 71, 2970–2978. [Google Scholar] [CrossRef] [Green Version]
- Vescovo, M.; Morelli, L.; Bottazzi, V.; Gasson, M.J. Conjugal Transfer of Broad-Host-Range Plasmid pAMbeta1 into Enteric Species of Lactic Acid Bacteria. Appl. Environ. Microbiol. 1983, 46, 753–755. [Google Scholar] [CrossRef] [Green Version]
- van der Lelie, D.; Venema, G. Bacillus subtilis generates a major specific deletion in pAMβ1. Appl. Environ. Microbiol. 1987, 53, 2458–2463. [Google Scholar] [CrossRef] [Green Version]
- Beutin, L.; Achtman, M. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J. Bacteriol. 1979, 139, 730–737. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.Y.; Mooney, R.A.; Nedialkov, Y.; Saba, J.; Mishanina, T.V.; Artsimovitch, I.; Landick, R.; Darst, S.A. Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell 2018, 173, 1650–1662. [Google Scholar] [CrossRef] [Green Version]
- Koraimann, G. Spread and Persistence of Virulence and Antibiotic Resistance Genes: A Ride on the F Plasmid Conjugation Module. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, M.J.; Hughes, C.; Koronakis, V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 1997, 26, 845–851. [Google Scholar] [CrossRef] [PubMed]




| Strains | Description and Genotype | Source or Reference |
|---|---|---|
| Escherichia coli | ||
| XL1-Blue | Used for regular cloning. endA1 gyrA96 (nalR) thi-1 recA1 relA1 lac glnV44 F’[Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17 (rK- mK+) | Laboratory stock (Stratagene) |
| Bacillus subtilis | ||
| 168 (1A700) | trpC2 | BGSC 1 |
| AND101 | B. subtilis 168 transformed with pAND101. trpC2, amyE:Pspank-gfp (SpecR) | [30] |
| AND127 | B. subtilis 168 transformed with pAND101. trpC2, amyE:Pspank-[Ter30pLS20]-gfp (SpecR) | [30] |
| EEF7 | B. subtilis 168 transformed with pEEF7. trpC2, amyE:Pspank-[Ter1pIP501]-gfp (SpecR) | This work |
| Plasmids | Description | Source or Reference |
| pEEF7 | pAND128 derivative containing terminator Ter1. Cloned fragment was generated by hybridization of primers oTer501_1A (SalI) and oTer501_1B (NheI). (AmpR) and (SpecR). | This work |
| Oligonucleotides | Sequence | Description |
| oTer501_1A | tcgacGTAACGTCTGTTTATGCAGATGAATTTCACTTTTTATTGAAG | Hybridization primer for generating TerInc18 fragment. Used in combination with oTer501_1B. SalI restriction site extension at the 5´end |
| oTer501_1B | ctagCTTCAATAAAAAGTGAAATTCATCTGCATAAACAGACGTTACg | Hybridization primer for generating TerInc18 fragment. Used in combination with oTer501_1A. NheI restriction site extension at the 5’ end |
| pDR111_U_sec | TGACTTTATCTACAAGGTGTGGC | Forward primer for verifying sequences of PCR fragments cloned into pAND101. |
| Name | Plasmid | Position * | Sequence (5′-3′) † |
|---|---|---|---|
| Ter1 | pIP501 | 5076 | GCAAATTGTAACGTCTGTTTATGCAGATGaaTTTCACTTTTTA |
| pAMβ1 | 5076 | GCAAATTGTAACGTCTGTTTATGCAGATGaaTTTCACTTTTTA | |
| pRE25 | 5076 | GCAAATTGTAACGTCTGTTTATGCAGATGaaTTTCACTTTTTA | |
| Ter2 | pIP501 | 14.848 | GTATTTATAAAAGCATGGTCGCAAGTTTCACTAGCAGCCATGCTTTTATTGAATC |
| pAMβ1 | 14.853 | GTATTTTTAAAAGCATGGTCGCAAGTTTCACTAGCAGCCATGCTTTTTTTGAATC | |
| pRE25 | 14.792 | GTAAATTTAAAAGCATGGTCGCAAGTTTCACTAGCAGCCATGCTTTTTTTGAATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miguel-Arribas, A.; Wu, L.J.; Michaelis, C.; Yoshida, K.-i.; Grohmann, E.; Meijer, W.J.J. Conjugation Operons in Gram-Positive Bacteria with and without Antitermination Systems. Microorganisms 2022, 10, 587. https://doi.org/10.3390/microorganisms10030587
Miguel-Arribas A, Wu LJ, Michaelis C, Yoshida K-i, Grohmann E, Meijer WJJ. Conjugation Operons in Gram-Positive Bacteria with and without Antitermination Systems. Microorganisms. 2022; 10(3):587. https://doi.org/10.3390/microorganisms10030587
Chicago/Turabian StyleMiguel-Arribas, Andrés, Ling Juan Wu, Claudia Michaelis, Ken-ichi Yoshida, Elisabeth Grohmann, and Wilfried J. J. Meijer. 2022. "Conjugation Operons in Gram-Positive Bacteria with and without Antitermination Systems" Microorganisms 10, no. 3: 587. https://doi.org/10.3390/microorganisms10030587
APA StyleMiguel-Arribas, A., Wu, L. J., Michaelis, C., Yoshida, K. -i., Grohmann, E., & Meijer, W. J. J. (2022). Conjugation Operons in Gram-Positive Bacteria with and without Antitermination Systems. Microorganisms, 10(3), 587. https://doi.org/10.3390/microorganisms10030587