Mapping Bacterial Biofilm on Features of Orthopedic Implants In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria
2.2. Implants
- Total Knee System (knee)
- Sliding Hip Screw (ss)
- 5.5 in by 0.5 in Fixation Plate (fix (5))
- 3.25 in by 0.4 in Fixation Plate (fix (3))
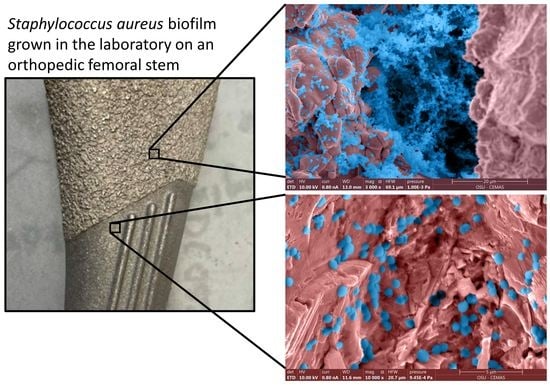
- Femoral Hip Stem (fhs)
- Hip Replacement Ball Joint (bjh)
- Modular Stem (mod)
- 7.3 MM TI cannulated screw 32 MM thread/45 MM (screw)
2.3. Growing Biofilm on Implants
2.4. IVIS Imaging
2.5. Scanning Electron Microscopy (SEM)
2.6. Roughness Characterization
2.7. Cleaning Explants
2.8. Quantification
2.9. Statistics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cataldo, M.A.; Petrosillo, N.; Cipriani, M.; Cauda, R.; Tacconelli, E. Prosthetic joint infection: Recent developments in diagnosis and management. J. Infect. 2010, 61, 443–448. [Google Scholar] [CrossRef]
- Johannsson, B.; Taylor, J.; Clark, C.R.; Shamsuddin, H.; Beekmann, S.E.; Polgreen, P.; Infectious Diseases Society of America Emerging Infections Network. Treatment approaches to prosthetic joint infections: Results of an Emerging Infections Network survey. Diagn. Microbiol. Infect. Dis. 2010, 66, 16–23. [Google Scholar] [CrossRef]
- Stoodley, P.; Kathju, S.; Hu, F.Z.; Erdos, G.; Levenson, J.E.; Mehta, N.; Dice, B.; Johnson, S.; Hall-Stoodley, L.; Nistico, L. Molecular and imaging techniques for bacterial biofilms in joint arthroplasty infections. Clin. Orthop. Relat. Res. 2005, 437, 31–40. [Google Scholar] [CrossRef]
- Darouiche, R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004, 350, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Halpern, M.; Kurtz, S.; Lau, E.; Mowat, F.; Ong, K. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J. Bone Surg. 2007, 89, 780–785. [Google Scholar]
- Gundtoft, P.H.; Pedersen, A.B.; Schønheyder, H.C.; Møller, J.K.; Overgaard, S. One-year incidence of prosthetic joint infection in total hip arthroplasty: A cohort study with linkage of the Danish Hip Arthroplasty Register and Danish Microbiology Databases. Osteoarthr. Cartil. 2017, 25, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Triffault-Fillit, C.; Ferry, T.; Laurent, F.; Pradat, P.; Dupieux, C.; Conrad, A.; Becker, A.; Lustig, S.; Fessy, M.-H.; Chidiac, C. Microbiologic epidemiology depending on time to occurrence of prosthetic joint infection: A prospective cohort study. Clin. Microbiol. Infect. 2019, 25, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.-D.; Wang, Y.-P.; Chen, C.-F.; Chen, H.-P. The incidence rate, trend and microbiological aetiology of prosthetic joint infection after total knee arthroplasty: A 13 years’ experience from a tertiary medical center in Taiwan. J. Microbiol. Immunol. Infect. 2018, 51, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev. Anti-Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef] [Green Version]
- McConoughey, S.J.; Howlin, R.; Granger, J.F.; Manring, M.M.; Calhoun, J.H.; Shirtliff, M.; Kathju, S.; Stoodley, P. Biofilms in periprosthetic orthopedic infections. Future Microbiol. 2014, 9, 987–1007. [Google Scholar] [CrossRef] [Green Version]
- Pestrak, M.J.; Gupta, T.T.; Dusane, D.H.; Guzior, D.V.; Staats, A.; Harro, J.; Horswill, A.R.; Stoodley, P. Investigation of synovial fluid induced Staphylococcus aureus aggregate development and its impact on surface attachment and biofilm formation. PLoS ONE 2020, 15, e0231791. [Google Scholar]
- Ricciardi, B.F.; Muthukrishnan, G.; Masters, E.; Ninomiya, M.; Lee, C.C.; Schwarz, E.M. Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: Biofilm and beyond. Curr. Rev. Musculoskelet. Med. 2018, 11, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.; Lewandowski, Z.; Caldwell, D.; Korber, D.; Lappin-Scott, H. 1079 M. Microb. Biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.; Lovering, A.M.; Webb, J.C. The role of microbial biofilms in prosthetic joint infections. Acta Orthop. 2015, 86, 147–158. [Google Scholar]
- Barth, E.; Myrvik, Q.M.; Wagner, W.; Gristina, A.G. In vitro and in vivo comparative colonization of Staphylococcus aureus and Staphylococcus epidermidis on orthopaedic implant materials. Biomaterials 1989, 10, 325–328. [Google Scholar] [CrossRef]
- Oga, M.; Sugioka, Y.; Hobgood, C.; Gristina, A.; Myrvik, Q. Surgical biomaterials and differential colonization by Staphylococcus epidermidis. Biomaterials 1988, 9, 285–289. [Google Scholar] [CrossRef]
- Gupta, T.T.; Gupta, N.K.; Pestrak, M.J.; Dusane, D.H.; Harro, J.M.; Horswill, A.R.; Stoodley, P. Staphylococcus aureus aggregates on orthopedic materials under varying levels of shear stress. Appl. Environ. Microbiol. 2020, 86, e01234-20. [Google Scholar] [CrossRef]
- Amalou, H.; Negussie, A.H.; Ranjan, A.; Chow, L.; Xu, S.; Kroeger, C.; Neeman, Z.; O’Grady, N.P.; Wood, B.J. Electrically conductive catheter inhibits bacterial colonization. J. Vasc. Interv. Radiol. 2014, 25, 797–802. [Google Scholar] [CrossRef]
- Feng, W.; Geng, Z.; Li, Z.; Cui, Z.; Zhu, S.; Liang, Y.; Liu, Y.; Wang, R.; Yang, X. Controlled release behaviour and antibacterial effects of antibiotic-loaded titania nanotubes. Mater. Sci. Eng. C 2016, 62, 105–112. [Google Scholar] [CrossRef]
- Kuehl, R.; Brunetto, P.S.; Woischnig, A.-K.; Varisco, M.; Rajacic, Z.; Vosbeck, J.; Terracciano, L.; Fromm, K.M.; Khanna, N. Preventing implant-associated infections by silver coating. Antimicrob. Agents Chemother. 2016, 60, 2467–2475. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, W.; Silva, P.; Silva, R.; Silva, G.; Machado, G.; Coelho, L.; Correia, M. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 2018, 98, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Bernthal, N.M.; Stavrakis, A.I.; Billi, F.; Cho, J.S.; Kremen, T.J.; Simon, S.I.; Cheung, A.L.; Finerman, G.A.; Lieberman, J.R.; Adams, J.S. A mouse model of post-arthroplasty Staphylococcus aureus joint infection to evaluate in vivo the efficacy of antimicrobial implant coatings. PLoS ONE 2010, 5, e12580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luca, A.; Gallazzi, E.; De Vecchi, E.; Brayda-Bruno, M.; Lovi, A.; Babbi, L.; Peretti, G.M.; Bidossi, A. Bacterial adhesion on spinal implants: An in vitro study of “hot spots”. J. Orthop. Res. 2021, 39, 2209–2216. [Google Scholar] [CrossRef]
- Plaut, R.D.; Mocca, C.P.; Prabhakara, R.; Merkel, T.J.; Stibitz, S. Stably luminescent Staphylococcus aureus clinical strains for use in bioluminescent imaging. PLoS ONE 2013, 8, e59232. [Google Scholar] [CrossRef] [PubMed]
- Goeres, D.M.; Loetterle, L.R.; Hamilton, M.A.; Murga, R.; Kirby, D.W.; Donlan, R.M. Statistical assessment of a laboratory method for growing biofilms. Microbiology 2005, 151, 757–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HORIBA Scientific. Determining Compounds in Cleaning Products with Raman Mapping. AZoM. 2020. Available online: https://www.azom.com/article.aspx?ArticleID=16576 (accessed on 21 February 2022).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Arnold, J.; Bailey, G. Surface finishes on stainless steel reduce bacterial attachment and early biofilm formation: Scanning electron and atomic force microscopy study. Poult. Sci. 2000, 79, 1839–1845. [Google Scholar] [CrossRef]
- James, G.A.; Boegli, L.; Hancock, J.; Bowersock, L.; Parker, A.; Kinney, B.M. Bacterial adhesion and biofilm formation on textured breast implant shell materials. Aesthetic Plast. Surg. 2019, 43, 490–497. [Google Scholar] [CrossRef] [Green Version]
- Jeong, W.-S.; Kwon, J.-S.; Lee, J.-H.; Uhm, S.-H.; Choi, E.H.; Kim, K.-M. Bacterial attachment on titanium surfaces is dependent on topography and chemical changes induced by nonthermal atmospheric pressure plasma. Biomed. Mater. 2017, 12, 045015. [Google Scholar] [CrossRef]
- Scardino, A.; Guenther, J.; De Nys, R. Attachment point theory revisited: The fouling response to a microtextured matrix. Biofouling 2008, 24, 45–53. [Google Scholar] [CrossRef]
- Sierra, D.R.; Edwardson, S.; Dearden, G. Laser surface texturing of titanium with thermal post-processing for improved wettability properties. Procedia CIRP 2018, 74, 362–366. [Google Scholar] [CrossRef]
- Scardino, A.; Harvey, E.; De Nys, R. Testing attachment point theory: Diatom attachment on microtextured polyimide biomimics. Biofouling 2006, 22, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.W.; Guillot, S.J.; Redick, J.A.; Browne, J.A. Removed antibiotic-impregnated cement spacers in two-stage revision joint arthroplasty do not show biofilm formation in vivo. J. Arthroplast. 2012, 27, 1796–1799. [Google Scholar] [CrossRef] [PubMed]
- Cowle, M.; Babatunde, A.; Rauen, W.; Bockelmann-Evans, B.N.; Barton, A. Biofilm development in water distribution and drainage systems: Dynamics and implications for hydraulic efficiency. Environ. Technol. Rev. 2014, 3, 31–47. [Google Scholar] [CrossRef]
- Stoodley, P.; Nistico, L.; Johnson, S.; Lasko, L.-A.; Baratz, M.; Gahlot, V.; Ehrlich, G.D.; Kathju, S. Direct demonstration of viable Staphylococcus aureus biofilms in an infected total joint arthroplasty: A case report. J. Bone Jt. Surg. 2008, 90, 1751. [Google Scholar] [CrossRef] [Green Version]
- Stoodley, P.; Conti, S.F.; DeMeo, P.J.; Nistico, L.; Melton-Kreft, R.; Johnson, S.; Darabi, A.; Ehrlich, G.D.; Costerton, J.W.; Kathju, S. Characterization of a mixed MRSA/MRSE biofilm in an explanted total ankle arthroplasty. FEMS Immunol. Med. Microbiol. 2011, 62, 66–74. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moore, K.; Gupta, N.; Gupta, T.T.; Patel, K.; Brooks, J.R.; Sullivan, A.; Litsky, A.S.; Stoodley, P. Mapping Bacterial Biofilm on Features of Orthopedic Implants In Vitro. Microorganisms 2022, 10, 586. https://doi.org/10.3390/microorganisms10030586
Moore K, Gupta N, Gupta TT, Patel K, Brooks JR, Sullivan A, Litsky AS, Stoodley P. Mapping Bacterial Biofilm on Features of Orthopedic Implants In Vitro. Microorganisms. 2022; 10(3):586. https://doi.org/10.3390/microorganisms10030586
Chicago/Turabian StyleMoore, Kelly, Niraj Gupta, Tripti Thapa Gupta, Khushi Patel, Jacob R. Brooks, Anne Sullivan, Alan S. Litsky, and Paul Stoodley. 2022. "Mapping Bacterial Biofilm on Features of Orthopedic Implants In Vitro" Microorganisms 10, no. 3: 586. https://doi.org/10.3390/microorganisms10030586
APA StyleMoore, K., Gupta, N., Gupta, T. T., Patel, K., Brooks, J. R., Sullivan, A., Litsky, A. S., & Stoodley, P. (2022). Mapping Bacterial Biofilm on Features of Orthopedic Implants In Vitro. Microorganisms, 10(3), 586. https://doi.org/10.3390/microorganisms10030586