Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community
Abstract
:1. Introduction
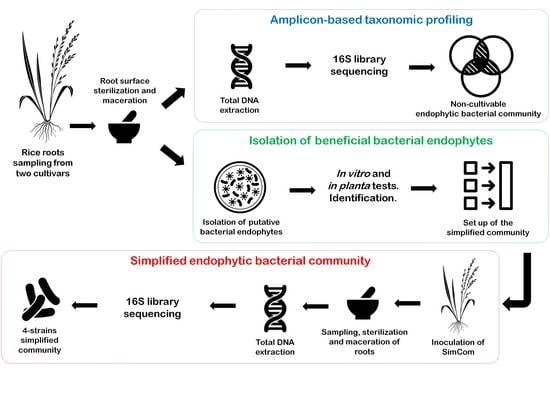
2. Materials and Methods
2.1. Sample Collection and Isolation of Bacteria
2.2. Total Bacterial Diversity of Rhizosphere and Endorhizosphere
2.3. Plant-Growth Promoting Traits
2.4. Identification of Selected Isolates
2.5. Seedling Early Growth, Endophytism and Plant-Growth Promotion Assay
2.6. Simplified Community Colonization Assay
2.7. Analyses of Sequencing Data
3. Results
3.1. Bacterial Diversity of Venezuelan Rice Rhizosphere and Endorhizosphere
3.2. Isolation of Culturable Bacteria from Rhizosphere and Endorhizosphere
3.3. Production of Indoleacetic Acid (IAA)
3.4. Molecular Identification
3.5. In Vitro Assays of Plant Beneficial Traits
3.6. Seedling Early Growth, Endophytism Assay and Plant-Growth Promotion
3.7. Simplified Community Inoculation, Colonization and Plant Growth Promotion
4. Discussion
4.1. Amplicon-Based Taxonomic Profiling
4.2. Isolation of Putative Endophytic Bacteria, Determination of Its PGP Traits and Plant Colonization
4.3. Seedling Inoculation with a Simplified Bacterial Community
4.4. Limitations of This Study
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAO Statistical Yearbook 2013: World Food and Agriculture; FAO: Rome, Italy, 2013; 307p. [Google Scholar]
- Naher, U.A.; Othman, R.; Panhwar, Q.A.; Ismail, M.R. Biofertilizer for sustainable rice production and reduction of environmental pollution. In Crop Production and Global Environmental Issues; Springer: Berlin, Germany, 2015; pp. 283–291. [Google Scholar]
- Singh, J.S.; Pandey, V.C.; Singh, D.P. Efficient soil microorganisms: A new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 2011, 140, 339–353. [Google Scholar] [CrossRef]
- Kim, Y.C.; Leveau, J.; Gardener, B.B.M.; Pierson, E.A.; Pierson, L.S.; Ryu, C.M. The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 2011, 77, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.N.; Padmavathy, S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffard, M.L.; Touraine, B.; Moenne-Loccoz, Y.; Muller, D.; Leqendre, L.; Wisniewski-Dye, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Alikhani, H.A.; Hosseini, H.M. Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2015, 2, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Zdor, R.E. Bacterial cyanogenesis: Impact on biotic interactions. J. Appl. Microbiol. 2015, 118, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; Mdel, C.O.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Mano, H.; Morisaki, H. Endophytic bacteria in the rice plant. Microbes Environ. 2008, 23, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Hardoin, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Bertani, I.; Abbruscato, P.; Piffanelli, P.; Subramoni, S.; Venturi, V. Rice bacterial endophytes: Isolation of a collection, identification of beneficial strains and microbiome analysis. Environ. Microbiol. Rep. 2016, 8, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marella, S. Bacterial endophytes in sustainable crop production: Applications, recent developments and challenges ahead. Int. J. Life Sci. Res. 2014, 2, 46–56. [Google Scholar]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Arenz, B.E.; Schlatter, D.C.; Bradeen, J.M.; Kinkel, L.L. Blocking primers reduce co-amplification of plant DNA when studying bacterial endophyte communities. J. Microbiol. Methods 2015, 117, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [PubMed]
- Pikovskaya, R. Mobilization of phosphorous in soil in connection with vital activity of some microbial species. Microbiologiya 1948, 17, 362–370. [Google Scholar]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Tatlor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Kohler, T.; Curty, L.K.; Barja, F.; van Delden, C.; Pechere, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [PubMed]
- Zlosnik, J.E.A.; Hird, T.J.; Fraenkel, M.C.; Moreira, L.M.; Henry, D.A.; Speert, D.P. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 2008, 46, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.S.; Hart, M.E.; Iandolo, J.J. Quantitative spectrophotometric assay for staphylococcal lipase. Appl. Environ. Microbiol. 1992, 58, 2815–2819. [Google Scholar] [PubMed]
- Huber, B.; Riedel, K.; Hentzer, M.; Heydorn, A.; Gotschlich, A.; Givskov, M.; Molin, S.; Eberl, L. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2001, 147, 2517–2528. [Google Scholar] [CrossRef] [PubMed]
- Mehnaz, S.; Baig, D.N.; Lazarovits, G. Genetic and Phenotypic Diversity of Plant Growth Promoting Rhizobacteria Isolated from Sugarcane Plants Growing in Pakistan. J. Microbiol. Biotechnol. 2010, 20, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [PubMed]
- Dereeper, A.; Guiqnon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Ledort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, R.; Pal, K.K.; Bhatt, D.M.; Chauhan, S.M. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004, 159, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Lingua, G.; Berta, G.; Lemanceau, P. Methods for studying root colonization by introduced beneficial bacteria. In Sustainable Agriculture, 2009; Springer: New York, NY, USA, 2009; pp. 601–615. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; College of Agriculture, University of California: Berkeley, CA, USA, 1950. [Google Scholar]
- Angiuoli, S.V.; Matalka, M.; Gussman, A.; Galens, K.; Vanqala, M.; Riley, D.R.; Arze, C.; White, J.R.; White, O.; Fricke, W.F. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinform. 2011, 12, 356. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombauqh, J.; Bittinqer, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robins, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Jost, L.; Devries, P.; Walla, T.; Greeney, H.; Chao, A.; Ricotta, C. Partitioning diversity for conservation analyses. Divers. Distrib. 2010, 16, 65–76. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Soman, C.; Waqner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Danql, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e20011793. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Dumont, M.G.; Yuan, Q.; Conrad, R. Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl. Environ. Microbiol. 2015, 81, 2244–2253. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Andreote, F.D.; Reinhold-Hurek, B.; Sessitsch, A.; van Overbeek, L.S.; van Elsas, J.D. Rice root-associated bacteria: Insights into community structures across10 cultivars. FEMS Microbiol. Ecol. 2011, 77, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 2017, 8, e00764–e007617. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Qiu, F.; Zhang, X.; Dai, X.; Dong, X.; Song, W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb. Ecol. 2008, 55, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The Plant Microbiota: Systems-Level Insights and Perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef] [PubMed]
- Sukumar, P.; Legué, V.; Vayssières, A.; Martin, F.; Tuskan, G.A.; Kalluri, U.C. Involvement of auxin pathways in modulating root architecture during beneficial plant-microorganism interactions. Plant Cell Environ. 2013, 36, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Ratering, S.; Suarez, C.; Montoya, A.M.Z.; Geissler-Plaum, R.; Schnell, S. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol. Res. 2015, 181, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, U.; Chakraborty, B.N.; Chakraborty, A.P. Influence of Serratia marcescens TRS-1 on growth promotion and induction of resistance in Camellia sinensis against Fomes lamaoensis. J. Plant Interact. 2010, 5, 261–272. [Google Scholar] [CrossRef]
- Neupane, S.; Hoqberq, N.; Alstrom, S.; Lucass, S.; Han, J.; Lapidus, A.; Chenq, J.F.; Bruce, D.; Goodqin, L.; Pitluck, S.; et al. Complete genome sequence of the rapeseed plant-growth promoting Serratia plymuthica strain AS9. Stand. Genom. Sci. 2012, 6, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Devi, U.; Khatri, I.; Kuamr, N.; Kumar, L.; Sharma, D.; Subramanian, S.; Saini, A.K. Draft Genome Sequence of a Plant Growth-Promoting Rhizobacterium, Serratia fonticola Strain AU-P3(3). Genome Announc. 2013, 1, e00946-13. [Google Scholar] [CrossRef] [PubMed]
- Prakamhang, J.; Minamisawa, K.; Teamtaisong, K.; Boonkerd, N.; Teaumroong, N. The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.). Appl. Soil Ecol. 2009, 42, 141–149. [Google Scholar] [CrossRef]
- Kaga, H.; Mano, H.; Tanaka, F.; Watanabe, A.; Kaneko, S.; Morisaki, H. Rice Seeds as Sources of Endophytic Bacteria. Microbes Environ. 2009, 24, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Banik, A.; Mukhopadhaya, S.K.; Dangar, T.K. Characterization of N2-fixing plant growth promoting endophytic and epiphytic bacterial community of Indian cultivated and wild rice (Oryza spp.) genotypes. Planta 2016, 243, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cristobal, J.; Garcia-Villaraco, A.; Ramos, B.; Gutierrez-Mañero, J.; Lucas, J.A. Priming of pathogenesis related-proteins and enzymes related to oxidative stress by plant growth promoting rhizobacteria on rice plants upon abiotic and biotic stress challenge. J. Plant Physiol. 2015, 188, 72–79. [Google Scholar] [CrossRef] [PubMed]
- De Roy, K.; Marzorati, M.; van den Abbeele, P.; Van de Wiele, T.; Boon, N. Synthetic microbial ecosystems: An exciting tool to understand and apply microbial communities. Environ. Microbiol. 2014, 16, 1472–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Muller, D.B.; Srinivas, G.; Garrido-Otyer, R.; Pptthoff, E.; Rott, M.; Dombrowski, N.; Munch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [PubMed]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 12, 12151. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Singh, A.; Singh, H.B.; Sarma, B.K. Plant genotype, microbial recruitment and nutritional security. Front. Plant Sci. 2015, 6, 608. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed]






| Samples | Plant Derived Reads | Bacterial Derived Reads | Average Length (bp) | |||
|---|---|---|---|---|---|---|
| # | % | # | % | |||
| (A) Amplicon-based taxonomic profiling | ||||||
| Pionero FL 2010 | Rhizospheres | 320 | 0.16 | 175,530 | 99.84 | 248 |
| Endorhizospheres | 60 | 0.06 | 81,171 | 99.94 | 247 | |
| DANAC SD20A | Rhizospheres | 81 | 0.09 | 49,374 | 99.91 | 249 |
| Endorhizospheres | 16 | 0.04 | 20,421 | 99.96 | 248 | |
| (B) Simplified community | ||||||
| Control endorhizospheres | 10 days | 94,362 | 53.16 | 83,143 | 46.84 | 247 |
| 20 days | 111,238 | 95.63 | 5083 | 4.37 | 247 | |
| 30 days | 153,205 | 98.41 | 2475 | 1.59 | 248 | |
| Inoculated endorhizospheres | 10 days | 3530 | 2.29 | 150,625 | 97.71 | 246 |
| 20 days | 100,128 | 57.54 | 73,887 | 42.46 | 248 | |
| 30 days | 300,845 | 91.34 | 28,523 | 8.66 | 248 | |
| Richness Estimator | Diversity Estimator | ||||||
|---|---|---|---|---|---|---|---|
| OTUs | Chao | ACE | Simpson | Shannon | ENS | ||
| Pionero FL2010 | R | 1497 | 1549.6 | 1586.8 | 0.078 | 4.28 | 72 |
| E | 794 | 825.5 | 855.2 | 0.089 | 3.74 | 42 | |
| SD20A | R | 1620 | 1663.4 | 1706.7 | 0.014 | 5.52 | 250 |
| E | 562 | 635.6 | 651.2 | 0.148 | 3.06 | 21 | |
| Rice Cultivar | Bacterial Isolate | Accession NCBI A | AccessionCVCM B | Closest Type Strain C | Reference Sequence D | Similarity |
|---|---|---|---|---|---|---|
| (%) | ||||||
| Pionero 2010 FL | E1101 | KY867521 | CVCM2317 | Bacillus velezensis CR-502(T) | AY603658 | 99.86 |
| E1103 | KY867522 | CVCM2318 | Pseudomonas gessardii DSM 17152(T) | MNPU01000117 | 99.93 | |
| E1108 | KY867523 | CVCM2319 | Pseudomonas chengduensis MBR(T) | EU307111 | 99.93 | |
| E1201 | KY867525 | CVCM2322 | Pseudomonas chengduensis MBR(T) | EU307111 | 99.59 | |
| E1205 | KY867526 | CVCM2324 | Pseudomonas oleovorans subsp. oleovorans DSM 1045(T) | NIUB01000072 | 99.25 | |
| E1308 | KY867527 | CVCM2326 | Pseudomonas gessardii DSM 17152(T) | MNPU01000117 | 99.93 | |
| DANAC SD20A | E2102 | KY867528 | CVCM2328 | Aeromonas veronii CECT 4257(T) | CDDK01000015 | 99.86 |
| E2105 | KY867529 | CVCM2329 | Serratia glossinae C1(T) | FJ790328 | 99.66 | |
| E2202 | KY867530 | - | Aeromonas hydrophila subsp. hydrophila ATCC 7966(T) | CP000462 | 99.93 | |
| E2205 | KY867531 | CVCM2330 | Aeromonas veronii CECT 4257(T) | CDDK01000015 | 100 | |
| E2309 | KY867532 | CVCM2331 | Serratia glossinae C1(T) | FJ790328 | 99.93 | |
| E2315 | KY867533 | CVCM2334 | Bacillus altitudinis 41KF2b(T) | ASJC01000029 | 99.93 | |
| E2321 | KY867534 | CVCM2335 | Rhizobium oryziradicis N19(T) | KX129901 | 98.08 | |
| E2330 | KY867535 | - | Delftia lacustris LMG 24775(T) | jgi.1102360 | 99.93 | |
| E2333 | KY867536 | CVCM2338 | Pseudomonas helmanticensis OHA11(T) | HG940537 | 99.31 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moronta-Barrios, F.; Gionechetti, F.; Pallavicini, A.; Marys, E.; Venturi, V. Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community. Microorganisms 2018, 6, 14. https://doi.org/10.3390/microorganisms6010014
Moronta-Barrios F, Gionechetti F, Pallavicini A, Marys E, Venturi V. Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community. Microorganisms. 2018; 6(1):14. https://doi.org/10.3390/microorganisms6010014
Chicago/Turabian StyleMoronta-Barrios, Felix, Fabrizia Gionechetti, Alberto Pallavicini, Edgloris Marys, and Vittorio Venturi. 2018. "Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community" Microorganisms 6, no. 1: 14. https://doi.org/10.3390/microorganisms6010014
APA StyleMoronta-Barrios, F., Gionechetti, F., Pallavicini, A., Marys, E., & Venturi, V. (2018). Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community. Microorganisms, 6(1), 14. https://doi.org/10.3390/microorganisms6010014