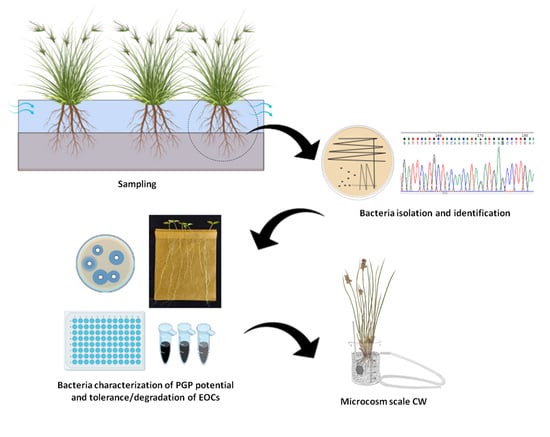
Root Bacteria Recruited by Phragmites australis in Constructed Wetlands Have the Potential to Enhance Azo-Dye Phytodepuration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacteria Collection Establishment
2.3. Bacteria Genotyping and Identification
2.4. In Vitro Screening of Plant Growth Promoting Activities and Rhizocompetence Potential
2.5. Plant Growth Promotion of Model Plants Under Liquid Substrate Culture and of Juncus acutus Plants in Soil
2.6. Bacterial Tolerance to Metals and Emerging Organic Pollutants (EOP)
2.7. In Vitro Study of Bacterial Dye Decolorization Potential
2.8. Bioaugmentation of Juncus acutus Microcosms for the Treatment of Mixed Contamination
3. Results
3.1. Cultivable Bacteria Associated to Phragmites australis in Constructed Wetlands Treating Municipal Wastewater
3.2. PGP Potential of P. australis Associated Bacteria
3.3. Metal and Organic Pollutant Tolerance and Degradation Potential of the Isolated Bacteria
3.4. Selection and Characterization of the Most Promising Strains for Bacterial Enhanced Phytodepuration
3.5. Evaluation of the Bacterial Contribution to Azo-Dye Removal by Juncus acutus in Microcosm-Scale CWs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jadhav, J.P.; Kalyani, D.C.; Telke, A.A.; Phugare, S.S.; Govindwar, S.P. Evaluation of the efficacy of a bacterial consortium for the removal of color, reduction of heavy metals, and toxicity from textile dye effluent. Bioresour. Technol. 2010, 101, 165–173. [Google Scholar] [CrossRef]
- Mahmood, F.; Shahid, M.; Hussain, S.; Shahzad, T.; Tahir, M.; Ijaz, M.; Hussain, A.; Mahmood, K.; Imran, M.; Babar, S.A.K. Potential plant growth-promoting strain Bacillus sp. SR-2-1/1 decolorized azo dyes through NADH-ubiquinone: Oxidoreductase activity. Bioresour. Technol. 2017, 235, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Shafqat, M.; Khalid, A.; Mahmood, T.; Siddique, M.T.; Han, J.I.; Habteselassie, M.Y. Evaluation of bacteria isolated from textile wastewater and rhizosphere to simultaneously degrade azo dyes and promote plant growth. J. Chem. Technol. Biotechnol. 2017, 92, 2760–2768. [Google Scholar] [CrossRef]
- Wang, Z.W.; Liang, J.S.; Liang, Y. Decolorization of Reactive Black 5 by a newly isolated bacterium Bacillus sp. YZU1. Int. Biodeterior. Biodegrad. 2013, 76, 41–48. [Google Scholar] [CrossRef]
- Lucas, M.S.; Amaral, C.; Sampaio, A.; Peres, J.A.; Dias, A.A. Biodegradation of the diazo dye Reactive Black 5 by a wild isolate of Candida oleophila. Enzym. Microb. Technol. 2006, 39, 51–55. [Google Scholar] [CrossRef]
- Khalid, A.; Arshad, M.; Crowley, D. Bioaugmentation of Azo Dyes. Biodegradation of Azo Dyes; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–37. [Google Scholar]
- Van der Zee, F.P.; Villaverde, S. Combined anaerobic–aerobic treatment of azo dyes—A short review of bioreactor studies. Water Res. 2005, 39, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.P. Azo Dye Removal Technologies. Austin J. Biotechnol. Bioeng. 2018, 5, 1090. [Google Scholar]
- Chaukura, N.; Gwenzi, W.; Tavengwa, N.; Manyuchi, M.M. Biosorbents for the removal of synthetic organics and emerging pollutants, opportunities and challenges for developing countries. Environ. Dev. 2016, 19, 84–89. [Google Scholar] [CrossRef]
- Frascari, D.; Zanaroli, G.; Motaleb, M.A.; Annen, G.; Belguith, K.; Borin, S.; Choukr-Allah, R.; Gibert, C.; Jaouani, A.; Kalogerakis, N.; et al. Integrated technological and management solutions for wastewater treatment and efficient agricultural reuse in Egypt, Morocco, and Tunisia. Integr. Environ. Assess. Manag. 2018, 14, 447–462. [Google Scholar] [CrossRef]
- Giorgetti, L.; Talouizte, H.; Merzouki, M.; Caltavuturo, L.; Geri, C.; Frassinetti, S. Genotoxicity evaluation of effluents from textile industries of the region Fez-Boulmane, Morocco: A case study. Ecotoxicol. Environ. Saf. 2011. [Google Scholar] [CrossRef]
- Garner, E.; Zhu, N.; Strom, L.; Edwards, M.; Pruden, A. A human exposome framework for guiding risk management and holistic assessment of recycled water quality. Environ. Sci. Water Res. Technol. 2016, 2, 580–598. [Google Scholar] [CrossRef] [Green Version]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Vymazal, J. Constructed wetlands for wastewater treatment. Water 2010, 2, 530–549. [Google Scholar] [CrossRef]
- Wang, J.; Tai, Y.; Man, Y.; Wang, R.; Feng, X.; Yang, Y.; Fan, J.; Guo, J.J.; Tao, R.; Yang, Y.; et al. Capacity of various single-stage constructed wetlands to treat domestic sewage under optimal temperature in Guangzhou City, South China. Ecol. Eng. 2018, 115, 35–44. [Google Scholar] [CrossRef]
- Wu, S.; Wallace, S.; Brix, H.; Kuschk, P.; Kirui, W.K.; Masi, F.; Dong, R. Treatment of industrial effluents in constructed wetlands: Challenges, operational strategies and overall performance. Environ. Pollut. 2015, 201, 107–120. [Google Scholar] [CrossRef]
- Gottschall, N.; Boutin, C.; Crolla, A.; Kinsley, C.; Champagne, P. The role of plants in the removal of nutrients at a constructed wetland treating agricultural (dairy) wastewater, Ontario, Canada. Ecol. Eng. 2007, 29, 154–163. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, G.; Ng, W.J.; Tan, S.K. A review on removing pharmaceutical contaminants from wastewater by constructed wetlands: Design, performance and mechanism. Sci. Total Environ. 2014, 468, 908–932. [Google Scholar] [CrossRef]
- Faulwetter, J.L.; Gagnon, V.; Sundberg, C.; Chazarenc, F.; Burr, M.D.; Brisson, J.; Camper, A.K.; Stein, O.R. Microbial processes influencing performance of treatment wetlands: A review. Ecol. Eng. 2009, 35, 987–1004. [Google Scholar] [CrossRef]
- Borruso, L.; Esposito, A.; Bani, A.; Ciccazzo, S.; Papa, M.; Zerbe, S.; Brusetti, L. Ecological diversity of sediment rhizobacteria associated with Phragmites australis along a drainage canal in the Yellow River watershed. J. Soils Sediments 2017, 17, 253–265. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473, 619–641. [Google Scholar] [CrossRef]
- Zhang, D.; Luo, J.; Lee, Z.M.P.; Maspolim, Y.; Gersberg, R.M.; Liu, Y.; Tan, S.K.; Ng, W.J. Characterization of bacterial communities in wetland mesocosms receiving pharmaceutical-enriched wastewater. Ecol. Eng. 2016, 90, 215–224. [Google Scholar] [CrossRef]
- Dimitroula, H.; Syranidou, E.; Manousaki, E.; Nikolaidis, N.P.; Karatzas, G.P.; Kalogerakis, N. Mitigation measures for chromium-VI contaminated groundwater—The role of endophytic bacteria in rhizofiltration. J. Hazard. Mater. 2015, 281, 114–120. [Google Scholar] [CrossRef]
- Tara, N.; Arslan, M.; Hussain, Z.; Iqbal, M.; Khan, Q.M.; Afzal, M. On-site performance of floating treatment wetland macrocosms augmented with dye-degrading bacteria for the remediation of textile industry wastewater. J. Clean. Prod. 2019, 217, 541–548. [Google Scholar] [CrossRef]
- Syranidou, E.; Christofilopoulos, S.; Politi, M.; Weyens, N.; Venieri, D.; Vangronsveld, J.; Kalogerakis, N. Bisphenol-A removal by the halophyte Juncus acutus in a phytoremediation pilot, Characterization and potential role of the endophytic community. J. Hazard. Mater. 2017, 323, 350–358. [Google Scholar] [CrossRef]
- Marasco, R.; Rolli, E.; Ettoumi, B.; Vigani, G.; Mapelli, F.; Borin, S.; Abou-Hadid, A.F.; El-Behairy, U.A.; Sorlini, C.; Cherif, A.; et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE 2012, 7, e48479. [Google Scholar] [CrossRef]
- Barac, T.; Taghavi, S.; Borremans, B.; Provoost, A.; Oeyen, L.; Colpaert, J.V.; Vangrosvelt, J.; Van Der Lelie, D. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 2004, 22, 583. [Google Scholar] [CrossRef]
- Vergani, L.; Mapelli, F.; Marasco, R.; Crotti, E.; Fusi, M.; Di Guardo, A.; Armiraglio, S.; Daffonchio, D.; Borin, S. Bacteria associated to plants naturally selected in a historical PCB polluted soil show potential to sustain natural attenuation. Front. Microbiol. 2017, 8, 1385. [Google Scholar] [CrossRef]
- Ferjani, R.; Marasco, R.; Rolli, E.; Cherif, H.; Cherif, A.; Gtari, M.; Boudabous, A.; Daffonchio, D.; Ouzari, H.I. The date palm tree rhizosphere is a niche for plant growth promoting bacteria in the oasis ecosystem. Biomed. Res. Int. 2015, 153851. [Google Scholar] [CrossRef]
- Mapelli, F.; Marasco, R.; Rolli, E.; Barbato, M.; Cherif, H.; Guesmi, A.; Ouzari, I.; Daffonchio, D.; Borin, S. Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. Biomed. Res. Int. 2013, 248078. [Google Scholar] [CrossRef]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. J. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.; Sørensen, J. Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiol. Ecol. 1997, 22, 183–192. [Google Scholar] [CrossRef]
- Santaella, C.; Schue, M.; Berge, O.; Heulin, T.; Achouak, W. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ. Microbiol. 2008, 10, 2150–2163. [Google Scholar] [CrossRef]
- Mi, Z.H.; Yu, Z.C.; Su, H.N.; Wang, L.; Chen, X.L.; Pang, X.; Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Zhou, B.C.; et al. Physiological and genetic analyses reveal a mechanistic insight into the multifaceted lifestyles of Pseudoalteromonas sp. SM 9913 adapted to the deep-sea sediment. Environ. Microbiol. 2015, 17, 3795–3806. [Google Scholar] [CrossRef]
- Burton, E.; Yakandawala, N.; LoVetri, K.; Madhyastha, M.S. A microplate spectrofluorometric assay for bacterial biofilms. J. Ind. Microbiol. Biotechnol. 2007, 34, 1–4. [Google Scholar] [CrossRef]
- Sharma, A.; Johri, B.N.; Sharma, A.K.; Glick, B.R. Plant growth-promoting bacterium Pseudomonas sp. strain GRP3 influences iron acquisition in mung bean (Vigna radiata L. Wilzeck). Soil Biol. Biochem. 2003, 35, 887–894. [Google Scholar] [CrossRef]
- Committee on Biological Agents (ABAS). Available online: https://www.baua.de/EN/Tasks/Committee-administration/ABAS/ABAS_node.html (accessed on 28 April 2019).
- Soussi, A.; Ferjani, R.; Marasco, R.; Guesmi, A.; Cherif, H.; Rolli, E.; Mapelli, F.; Ouzari, H.I.; Daffonchio, D.; Cherif, A. Plant-associated microbiomes in arid lands: Diversity, ecology and biotechnological potential. Plant Soil 2016. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-Aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots shaping their microbiome, global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Shehzadi, M.; Fatima, K.; Imran, A.; Mirza, M.S.; Khan, Q.M.; Afzal, M. Ecology of bacterial endophytes associated with wetland plants growing in textile effluent for pollutant-degradation and plant growth-promotion potentials. Plant. Biosyst. 2016, 150, 1261–1270. [Google Scholar] [CrossRef]
- Calheiros, C.S.C.; Pereira, S.I.A.; Brix, H.; Rangel, A.O.S.S.; Castro, P.M.L. Assessment of culturable bacterial endophytic communities colonizing Canna flaccida inhabiting a wastewater treatment constructed wetland. Ecol. Eng. 2017, 98, 418–426. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhu, J.N.; Zhai, Z.H.; Zhang, Q. Endophytic bacterial diversity in roots of Phragmites australis in constructed Beijing Cuihu Wetland (China). FEMS Microbiol. Lett. 2010, 309, 84–93. [Google Scholar] [CrossRef]
- Sauvêtre, A.; Schröder, P. Uptake of carbamazepine by rhizomes and endophytic bacteria of Phragmites australis. Front. Plant Sci. 2015, 6, 83. [Google Scholar] [CrossRef]
- Marasco, R.; Mapelli, F.; Rolli, E.; Mosqueira, M.J.; Fusi, M.; Bariselli, P.; Reddy, M.; Cherif, A.; Tsiami, G.; Borin, S.; et al. Salicornia strobilacea (synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front. Microbiol. 2016, 7, 1286. [Google Scholar] [CrossRef]
- Vergani, L.; Mapelli, F.; Zanardini, E.; Terzaghi, E.; Di Guardo, A.; Morosini, C.; Raspa, G.; Borin, S. Phyto-rhizoremediation of polychlorinated biphenyl contaminated soils: An outlook on plant-microbe beneficial interactions. Sci. Total Environ. 2017, 575, 1395–1406. [Google Scholar] [CrossRef]
- Borruso, L.; Bacci, G.; Mengoni, A.; De Philippis, R.; Brusetti, L. Rhizosphere effect and salinity competing to shape microbial communities in Phragmites australis (Cav.) Trin. ex-Steud. FEMS Microbiol. Lett. 2014, 359, 193–200. [Google Scholar] [CrossRef]
- Behera, P.; Mohapatra, M.; Adhya, T.K.; Suar, M.; Pattnaik, A.K.; Rastogi, G. Structural and metabolic diversity of rhizosphere microbial communities of Phragmites karka in a tropical coastal lagoon. Appl. Soil Ecol. 2018, 125, 202–212. [Google Scholar] [CrossRef]
- Abed, R.M.; Al-Kharusi, S.; Gkorezis, P.; Prigent, S.; Headley, T. Bacterial communities in the rhizosphere of Phragmites australis from an oil-polluted wetland. Arch. Agron. Soil Sci. 2018, 64, 360–370. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome, looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef]
- Wu, S.; Carvalho, P.N.; Müller, J.A.; Manoj, V.R.; Dong, R. Sanitation in constructed wetlands: A review on the removal of human pathogens and fecal indicators. Sci. Total Environ. 2016, 541, 8–22. [Google Scholar] [CrossRef]
- Rehman, K.; Imran, A.; Amin, I.; Afzal, M. Inoculation with bacteria in floating treatment wetlands positively modulates the phytoremediation of oil field wastewater. J. Hazard. Mater. 2018, 349, 242–251. [Google Scholar] [CrossRef]
- Ijaz, A.; Shabir, G.; Khan, Q.M.; Afzal, M. Enhanced remediation of sewage effluent by endophyte-assisted floating treatment wetlands. Ecol. Eng. 2015, 84, 58–66. [Google Scholar] [CrossRef]
- Fountoulakis, M.S.; Daskalakis, G.; Papadaki, A.; Kalogerakis, N.; Manios, T. Use of halophytes in pilot-scale horizontal flow constructed wetland treating domestic wastewater. Environ. Sci. Pollut. Res. 2017, 24, 16682–16689. [Google Scholar] [CrossRef]
- Navarro-Torre, S.; Mateos-Naranjo, E.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Isolation of plant-growth-promoting and metal-resistant cultivable bacteria from Arthrocnemum macrostachyum in the Odiel marshes with potential use in phytoremediation. Mar. Pollut. Bull. 2016, 110, 133–142. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Sa, T. Metal tolerating methylotrophic bacteria reduces nickel and cadmium toxicity and promotes plant growth of tomato (Lycopersicon esculentum L.). Chemosphere 2007, 69, 220–228. [Google Scholar] [CrossRef]
- Baoune, H.; El Hadj-Khelil, A.O.; Pucci, G.; Sineli, P.; Loucif, L.; Polti, M.A. Petroleum degradation by endophytic Streptomyces spp. isolated from plants grown in contaminated soil of southern Algeria. Ecotoxicol. Environ. Saf. 2018, 147, 602–609. [Google Scholar] [CrossRef]
- Mapelli, F.; Marasco, R.; Balloi, A.; Rolli, E.; Cappitelli, F.; Daffonchio, D.; Borin, S. Mineral–microbe interactions, biotechnological potential of bioweathering. J. Biotechnol. 2012, 157, 473–481. [Google Scholar] [CrossRef]
- Loos, R.; Hanke, G.; Umlauf, G.; Eisenreich, S.J. LC–MS–MS analysis and occurrence of octyl-and nonylphenol; their ethoxylates and their carboxylates in Belgian and Italian textile industry, waste water treatment plant effluents and surface waters. Chemosphere 2007, 66, 690–699. [Google Scholar] [CrossRef]
- Xue, J.; Liu, W.; Kannan, K. Bisphenols, benzophenones, and bisphenol A diglycidyl ethers in textiles and infant clothing. Environ. Sci. Technol. 2017, 51, 5279–5286. [Google Scholar] [CrossRef]
- Noreen, M.; Shahid, M.; Iqbal, M.; Nisar, J. Measurement of cytotoxicity and heavy metal load in drains water receiving textile effluents and drinking water in vicinity of drains. Measurement 2017, 109, 88–99. [Google Scholar] [CrossRef]
- Hussain, Z.; Arslan, M.; Malik, M.H.; Mohsin, M.; Iqbal, S.; Afzal, M. Integrated perspectives on the use of bacterial endophytes in horizontal flow constructed wetlands for the treatment of liquid textile effluent, phytoremediation advances in the field. J. Environ. Manag. 2018, 224, 387–395. [Google Scholar] [CrossRef]
- Rajkumar, M.; Sandhya, S.; Prasad, M.N.V.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef]
- Syranidou, E.; Christofilopoulos, S.; Gkavrou, G.; Thijs, S.; Weyens, N.; Vangronsveld, J.; Kalogerakis, N. Exploitation of endophytic bacteria to enhance the phytoremediation potential of the wetland helophyte Juncus acutus. Front. Microbiol. 2016, 7, 1016. [Google Scholar] [CrossRef]
- Sun, E.; Liu, S.; Hancock, R.E. Surfing motility, a conserved yet diverse adaptation among motile bacteria. J. Bacteriol. 2018. [Google Scholar] [CrossRef]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634. [Google Scholar] [CrossRef]
- Kunito, T.; Saeki, K.; Nagaoka, K.; Oyaizu, H.; Matsumoto, S. Characterization of copper-resistant bacterial community in rhizosphere of highly copper-contaminated soil. Eur. J. Soil Biol. 2001, 37, 95–102. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Mahfouz, N.; Caucci, S.; Achatz, E.; Semmler, T.; Guenther, S.; Berendonk, T.U.; Schroeder, M. High genomic diversity of multi-drug resistant wastewater Escherichia coli. Sci. Rep. 2018, 8, 8928. [Google Scholar] [CrossRef]
- Syranidou, E.; Christofilopoulos, S.; Kalogerakis, N. Juncus spp.—The helophyte for all (phyto) remediation purposes? N. Biotechnol. 2017, 38, 43–55. [Google Scholar] [CrossRef]
- Vymazal, J. Constructed wetlands for treatment of industrial wastewaters: A review. Ecol. Eng. 2014, 73, 724–751. [Google Scholar] [CrossRef]
- Davies, L.C.; Carias, C.C.; Novais, J.M.; Martins-Dias, S. Phytoremediation of textile effluents containing azo dye by using Phragmites australis in a vertical flow intermittent feeding constructed wetland. Ecol. Eng. 2005, 25, 594–605. [Google Scholar] [CrossRef]
- Mbuligwe, S.E. Comparative treatment of dye-rich wastewater in engineered wetland systems (EWSs) vegetated with different plants. Water Res. 2005, 39, 271–280. [Google Scholar] [CrossRef]
- Jager, D.; Kupka, D.; Vaclavikova, M.; Ivanicova, L.; Gallios, G. Degradation of Reactive Black 5 by electrochemical oxidation. Chemosphere 2018, 190, 405–416. [Google Scholar] [CrossRef]
- Zaidi, S.Z.J.; Harito, C.; Walsh, F.C.; de León, C.P. Decolourisation of reactive black-5 at an RVC substrate decorated with PbO2/TiO2 nanosheets prepared by anodic electrodeposition. J. Solid State Electr. 2018, 1–12. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, X.W.; Su, J.Q.; Tian, Y.; Xiong, X.J.; Zheng, T.L. Biological decolorization of the reactive dyes Reactive Black 5 by a novel isolated bacterial strain Enterobacter sp. EC3. J. Hazard. Mater. 2009, 171, 654–659. [Google Scholar] [CrossRef]
- Abdelwahab, R.A.I.; Cherif, A.; Cristina, C.R.U.Z.; Nabti, E. Extracts from Marine Macroalgae and Opuntia ficus-indica Cladodes Enhance Halotolerance and Enzymatic Potential of Diazotrophic Rhizobacteria and Their Impact on Wheat Germination Under Salt Stress. Pedosphere 2018, 28, 241–254. [Google Scholar]
- Liste, H.H.; Felgentreu, D. Crop growth, culturable bacteria, and degradation of petrol hydrocarbons (PHCs) in a long-term contaminated field soil. Appl. Soil Ecol. 2006, 31, 43–52. [Google Scholar] [CrossRef]
- Yousaf, S.; Afzal, M.; Reichenauer, T.G.; Brady, C.L.; Sessitsch, A. Hydrocarbon degradation, plant colonization and gene expression of alkane degradation genes by endophytic Enterobacter ludwigii strains. Environ. Pollut. 2011, 159, 2675–2683. [Google Scholar] [CrossRef]
- Shehzadi, M.; Afzal, M.; Khan, M.U.; Islam, E.; Mobin, A.; Anwar, S.; Khan, Q.M. Enhanced degradation of textile effluent in constructed wetland system using Typha domingensis and textile effluent-degrading endophytic bacteria. Water Res. 2014, 58, 152–159. [Google Scholar] [CrossRef]
- Gottlieb, A.; Shaw, C.; Smith, A.; Wheatley, A.; Forsythe, S. The toxicity of textile reactive azo dyes after hydrolysis and decolourisation. J. Biotechnol. 2003, 101, 49–56. [Google Scholar] [CrossRef]






| Strain | Closest Described Relative | IAA | ACC-d | Prot. | Hydroponic Experiment | Biofilm | Swimming | Swarming | EPS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Germin. | Root-l | Shoot-l | 2ary Roots | SVI | Root-dw | |||||||||
| CWMP-8R25 | Pseudomonas fluorescens | + | + | * | ** | ** | * | 44% | ||||||
| CWMP-8R34 | Microbacterium oxydans | + | + | ** | * | 84% | ||||||||
| CWMP-8R67 | Microbacterium maritypicum | + | + | + | *** | * | 52% | |||||||
| CWMP-8R71 | Flavobacterium johnsoniae | *** | *** | *** | 6% | |||||||||
| CWMP-8R75 | Lysinibacillus fusiformis | + | *** | 66% | + | + | ||||||||
| CWMP-8R78 | Enterobacter ludwigii | + | + | *** | *** | *** | 21% | + | ||||||
| Strain | Closest Described Relative | BPA | Antibiotic Resistance | Metal Tolerance (mM) | Decolorization | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | C | KF | TET | VA | RA | CdCl2 | NiCl2 | ZnCl2 | MIX1 | MIX2 | RB5 | BR | TB | BS2-G | |||||||||
| 0.05 | 0.5 | 1 | 0.5 | 1 | 2 | 0.5 | 1 | 2 | |||||||||||||||
| CWMP-8R25 | Pseudomonas fluorescens | + | R | R | R | + | + | + | + | + | + | + | + | + | + | + | 68% | 10% | 0% | 22% | |||
| CWMP-8R34 | Microbacterium oxydans | + | R | R | R | R | + | + | + | + | + | + | + | + | + | + | + | 71% | 20% | 7% | 25% | ||
| CWMP-8R67 | Microbacterium maritypicum | + | R | R | + | + | + | + | + | + | + | + | + | + | + | 64% | 21% | 4% | 13% | ||||
| CWMP-8R71 | Flavobacterium johnsoniae | + | R | R | R | R | + | + | + | + | + | + | + | + | + | 74% | 85% | 58% | 15% | ||||
| CWMP-8R75 | Lysinibacillus fusiformis | + | R | + | + | + | + | + | + | + | + | + | 74% | 39% | 29% | 15% | |||||||
| CWMP-8R78 | Enterobacter ludwigii | + | R | R | + | + | + | + | + | + | + | + | + | + | 27% | 22% | 35% | 45% | |||||
| Antibiotic Resistance (R) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Code of the Isolates | CIP | C | KF | TET | VA | RA | N of R | % of Isolates |
| 1 | E13 | R | R | R | R | R | 5 | 2 | |
| 2 | R34, R52 | R | R | R | R | 4 | 19 | ||
| 3 | E2, E8, E14, E28, E33, R32 | R | R | R | R | ||||
| 4 | R12 | R | R | R | R | ||||
| 5 | R71 | R | R | R | R | ||||
| 6 | R20 | R | R | R | R | ||||
| 7 | R25 | R | R | R | 3 | 19 | |||
| 8 | E16, E42, E73, R65, R76, R79 | R | R | R | |||||
| 9 | E6, E21, R4, R69 | R | R | R | |||||
| 10 | R3 | R | R | 2 | 14 | ||||
| 11 | R67 | R | R | ||||||
| 12 | R17, R78 | R | R | ||||||
| 13 | R6, R28 | R | R | ||||||
| 14 | R22, R26 | R | R | ||||||
| 15 | E15, R2, R16, R23, R33, R39, R49, R50, R64, R75, R80 | R | 1 | 22 | |||||
| 16 | R8, R19 | R | |||||||
| 17 | E27, R1, R7, R9, R10, R15, R31, R38, R40, R47, R57, R68, R72, R77 | 0 | 24 | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riva, V.; Mapelli, F.; Syranidou, E.; Crotti, E.; Choukrallah, R.; Kalogerakis, N.; Borin, S. Root Bacteria Recruited by Phragmites australis in Constructed Wetlands Have the Potential to Enhance Azo-Dye Phytodepuration. Microorganisms 2019, 7, 384. https://doi.org/10.3390/microorganisms7100384
Riva V, Mapelli F, Syranidou E, Crotti E, Choukrallah R, Kalogerakis N, Borin S. Root Bacteria Recruited by Phragmites australis in Constructed Wetlands Have the Potential to Enhance Azo-Dye Phytodepuration. Microorganisms. 2019; 7(10):384. https://doi.org/10.3390/microorganisms7100384
Chicago/Turabian StyleRiva, Valentina, Francesca Mapelli, Evdokia Syranidou, Elena Crotti, Redouane Choukrallah, Nicolas Kalogerakis, and Sara Borin. 2019. "Root Bacteria Recruited by Phragmites australis in Constructed Wetlands Have the Potential to Enhance Azo-Dye Phytodepuration" Microorganisms 7, no. 10: 384. https://doi.org/10.3390/microorganisms7100384
APA StyleRiva, V., Mapelli, F., Syranidou, E., Crotti, E., Choukrallah, R., Kalogerakis, N., & Borin, S. (2019). Root Bacteria Recruited by Phragmites australis in Constructed Wetlands Have the Potential to Enhance Azo-Dye Phytodepuration. Microorganisms, 7(10), 384. https://doi.org/10.3390/microorganisms7100384