Characteristics of a Colistin-Resistant Escherichia coli ST695 Harboring the Chromosomally-Encoded mcr-1 Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Antimicrobial Susceptibility Testing
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Bioinformatical Analysis
2.4. Plasmid Conjugation
3. Results
3.1. Antimicrobial Susceptibility Profile of E. coli HeN100
3.2. Genetic Basis for the Resistant Profile of E. coli HeN100
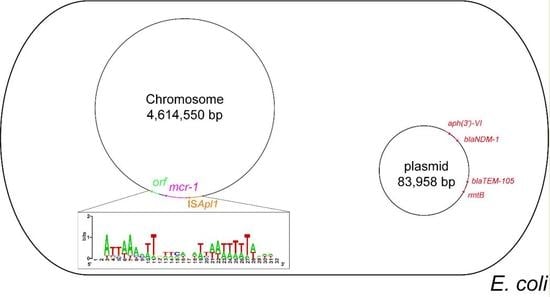
3.3. Genetic Environment of the Chromosomal mcr-1
3.4. Genetic Structure of the Blandm-1-Carrying Plasmid
3.5. The Transferability of the Blandm-1-Carrying Plasmid
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Trimble, M.J.; Mlynarcik, P.; Kolar, M.; Hancock, R.E. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist. Updates 2010, 13, 132–138. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Multidrug-Resistant Gram-Negative Pathogens: The Urgent Need for ‘Old’ Polymyxins. Adv. Exp. Med. Biol. 2019, 1145, 9–13. [Google Scholar] [CrossRef]
- Yao, X.; Doi, Y.; Zeng, L.; Lv, L.; Liu, J.H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016, 16, 288–289. [Google Scholar] [CrossRef]
- Li, Z.; Cao, Y.; Yi, L.; Liu, J.H.; Yang, Q. Emergent Polymyxin Resistance: End of an Era? Open Forum. Infect. Dis. 2019, 6. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 2019, 1145, 55–71. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Doumith, M.; Godbole, G.; Ashton, P.; Larkin, L.; Dallman, T.; Day, M.; Day, M.; Muller-Pebody, B.; Ellington, M.J.; de Pinna, E.; et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 2016, 71, 2300–2305. [Google Scholar] [CrossRef]
- Li, R.; Yu, H.; Xie, M.; Chen, K.; Dong, N.; Lin, D.; Chan, E.W.; Chen, S. Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 2018, 51, 578–585. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy in Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.; Zhang, Z.; Feng, J.; Kang, H.; Fang, L.; Jiang, X.; Zhang, D.; Zhan, Z.; Zhou, D.; et al. Structural genomics of pNDM-BTR harboring In191 and Tn6360, and other bla NDM-carrying IncN1 plasmids. Future Microbiol. 2017, 12, 1271–1281. [Google Scholar] [CrossRef]
- Lin, D.; Xie, M.; Li, R.; Chen, K.; Chan, E.W.; Chen, S. IncFII Conjugative Plasmid-Mediated Transmission of blaNDM-1 Elements among Animal-Borne Escherichia coli Strains. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Sun, J.; Yang, R.S.; Zhang, Q.; Feng, Y.; Fang, L.X.; Xia, J.; Li, L.; Lv, X.Y.; Duan, J.H.; Liao, X.P.; et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 2016, 1, 16176. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef]
- Abdul Momin, M.H.F.; Liakopoulos, A.; Bean, D.C.; Phee, L.M.; Wareham, D.W. A novel plasmid-mediated polymyxin resistance determinant (mcr-1.8) in Escherichia coli recovered from broiler chickens in Brunei Darussalam. J. Antimicrob. Chemother. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zajac, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Zietek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated from Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef] [PubMed]
- Martino, F.; Tijet, N.; Melano, R.; Petroni, A.; Heinz, E.; De Belder, D.; Faccone, D.; Rapoport, M.; Biondi, E.; Rodrigo, V.; et al. Isolation of five Enterobacteriaceae species harbouring blaNDM-1 and mcr-1 plasmids from a single paediatric patient. PLoS ONE 2019, 14, e0221960. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Li, X.; Hu, Z.; Li, Z.; Lv, Y.; Lei, M.; Wu, B.; Chen, H.; Wang, X. Characteristics of Carbapenem-Resistant and Colistin-Resistant Escherichia coli Co-Producing NDM-1 and MCR-1 from Pig Farms in China. Microorganisms 2019, 7, 482. [Google Scholar] [CrossRef] [Green Version]
- Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Canton, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 2008, 14, 195–200. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.H.; Andreasen, M.R.; Pedersen, M.S.; Westh, H.; Jelsbak, L.; Schonning, K. Resistance to piperacillin/tazobactam in Escherichia coli resulting from extensive IS26-associated gene amplification of blaTEM-1. J. Antimicrob. Chemother. 2019. [Google Scholar] [CrossRef]
- Aeksiri, N.; Toanan, W.; Sawikan, S.; Suwannarit, R.; Pungpomin, P.; Khieokhajonkhet, A.; Niumsup, P.R. First Detection and Genomic Insight into mcr-1 Encoding Plasmid-Mediated Colistin-Resistance Gene in Escherichia coli ST101 Isolated from the Migratory Bird Species Hirundo rustica in Thailand. Microb. Drug Resist. 2019. [Google Scholar] [CrossRef]
- Poirel, L.; Kieffer, N.; Nordmann, P. In Vitro Study of ISApl1-Mediated Mobilization of the Colistin Resistance Gene mcr-1. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Szabo, M.; Kiss, J.; Kotany, G.; Olasz, F. Importance of illegitimate recombination and transposition in IS30-associated excision events. Plasmid 1999, 42, 192–209. [Google Scholar] [CrossRef] [PubMed]
- Szabo, M.; Kiss, J.; Nagy, Z.; Chandler, M.; Olasz, F. Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. J. Mol. Biol. 2008, 375, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Tegetmeyer, H.E.; Fricke, K.; Baltes, N. An isogenic Actinobacillus pleuropneumoniae AasP mutant exhibits altered biofilm formation but retains virulence. Vet. Microbiol. 2009, 137, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-beta-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Antibiotics | MIC Values (μg/mL) and Interpretation | Interpretation Criteria (μg/mL) | |||
|---|---|---|---|---|---|
| HeN100 | ATCC25922 | S | I | R | |
| CL ǂ | >4 (R) | ≤1 (S) | ≤2 | - | >2 |
| AMK § | >32 (R) | ≤8 (S) | ≤16 | 32 | ≥64 |
| GEN § | >8 (R) | ≤2 (S) | ≤4 | 8 | ≥16 |
| TOB § | >8 (R) | ≤2 (S) | ≤4 | 8 | ≥16 |
| IPM § | >8 (R) | ≤0.25 (S) | ≤1 | 2 | ≥4 |
| MRP § | >8 (R) | ≤0.13 (S) | ≤1 | 2 | ≥4 |
| ETP § | >2 (R) | ≤0.25 (S) | ≤0.5 | 1 | ≥2 |
| CFZ § | >16 (R) | ≤2 (S) | ≤16 | - | ≥32 |
| CFX § | >16 (R) | ≤4 (S) | ≤8 | 16 | ≥32 |
| FOX § | >16 (R) | ≤4 (S) | ≤8 | 16 | ≥32 |
| CAZ § | >32 (R) | ≤1 (S) | ≤4 | 8 | ≥16 |
| CRO § | >32 (R) | ≤1 (S) | ≤1 | 2 | ≥4 |
| CPM § | >16 (R) | ≤1 (S) | ≤2 | - | ≥16 |
| AMC § | >32/16 (R) | ≤8/4 (S) | ≤8/4 | 16/8 | ≥32/16 |
| AMS § | >16/8 (R) | ≤4/2 (S) | ≤8/4 | 16/8 | ≥32/16 |
| PTZ § | >64/4 (R) | ≤4/4 (S) | ≤16/4 | 32/4–64/4 | ≥128/4 |
| CHL § | >16 (R) | ≤4 (S) | ≤8 | 16 | ≥32 |
| MXF ǂ | >2 (R) | ≤0.5 (S) | ≤0.25 | - | >0.25 |
| CIP § | 2 (R) | ≤0.5 (S) | ≤1 | 2 | ≥4 |
| LVX § | 2 (S) | ≤1 (S) | ≤2 | 4 | ≥8 |
| NOR § | 4 (S) | ≤2 (S) | ≤4 | 8 | ≥16 |
| TET § | >8 (R) | ≤2 (S) | ≤4 | 8 | ≥16 |
| MIN § | 16 (R) | ≤1 (S) | ≤4 | 8 | ≥16 |
| TGC ǂ | 2 | ≤1 (S) | ≤1 | - | >2 |
| NIT § | >64 (R) | ≤16 (S) | ≤32 | 64 | ≥128 |
| FOS § | ≤16 (S) | ≤16 (S) | ≤64 | 128 | ≥256 |
| SXT | ≤1/19 (S) | ≤1/19 (S) | ≤2/38 | - | ≥4/76 |
| AZM § | ≤2 (S) | ≤2 (S) | ≤4 | 8 | ≥16 |
| Antibiotics Tested | Minimum Inhibitory Concentration (μg/mL) | |
|---|---|---|
| Transconjugants | E. coli C600 | |
| Amikacin | >32 (R) | ≤8 (S) |
| Gentamicin | >8 (R) | ≤2 (S) |
| Tobramycin | >8 (R) | ≤2 (S) |
| Ertapenem | >2 (R) | ≤0.25 (S) |
| Imipenem | >8 (R) | 0.5 (S) |
| Meropenem | >8 (R) | ≤0.13 (S) |
| Cefazolin | >16 (R) | 4 (S) |
| Cefuroxime | >16 (R) | 16 (R) |
| Cefoxitin | >16 (R) | 8 (S) |
| Ceftazidime | >32 (R) | ≤1 (S) |
| Ceftriaxone | >32 (R) | ≤1 (S) |
| Cefepime | 16 (R) | ≤1 (S) |
| Amoxicillin/clavulanate | >32/16 (R) | ≤8/4 (S) |
| Ampicillin/sulbactam | >16/8 (R) | 8/4 (S) |
| Piperacillin/tazobactam | >64/4 (R) | ≤4/4 (S) |
| Colistin | ≤1 (S) | ≤1 (S) |
| Chloramphenicol | ≤4 (S) | ≤4 (S) |
| Moxifloxacin | ≤0.5 (S) | ≤0.5 (S) |
| Ciprofloxacin | ≤0.5 (S) | ≤0.5 (S) |
| Levofloxacin | ≤1 (S) | ≤1 (S) |
| Norfloxacin | ≤2 (S) | ≤2 (S) |
| Tetracycline | ≤2 (S) | ≤2 (S) |
| Minocycline | ≤1 (S) | ≤1 (S) |
| Trimethoprim/sulfamethoxazole | ≤1/19 (S) | ≤1/19 (S) |
| Aztreonam | ≤2 (S) | ≤2 (S) |
| Fosfomycin | ≤16 (S) | ≤16 (S) |
| Nitrofurantoin | ≤16 (S) | ≤16 (S) |
| Tigecycline | ≤1 (S) | ≤1 (S) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Hu, Z.; Li, Z.; Li, X.; Jia, C.; Zhang, X.; Wu, B.; Chen, H.; Wang, X. Characteristics of a Colistin-Resistant Escherichia coli ST695 Harboring the Chromosomally-Encoded mcr-1 Gene. Microorganisms 2019, 7, 558. https://doi.org/10.3390/microorganisms7110558
Peng Z, Hu Z, Li Z, Li X, Jia C, Zhang X, Wu B, Chen H, Wang X. Characteristics of a Colistin-Resistant Escherichia coli ST695 Harboring the Chromosomally-Encoded mcr-1 Gene. Microorganisms. 2019; 7(11):558. https://doi.org/10.3390/microorganisms7110558
Chicago/Turabian StylePeng, Zhong, Zizhe Hu, Zugang Li, Xiaosong Li, Chaoying Jia, Xiaoxue Zhang, Bin Wu, Huanchun Chen, and Xiangru Wang. 2019. "Characteristics of a Colistin-Resistant Escherichia coli ST695 Harboring the Chromosomally-Encoded mcr-1 Gene" Microorganisms 7, no. 11: 558. https://doi.org/10.3390/microorganisms7110558
APA StylePeng, Z., Hu, Z., Li, Z., Li, X., Jia, C., Zhang, X., Wu, B., Chen, H., & Wang, X. (2019). Characteristics of a Colistin-Resistant Escherichia coli ST695 Harboring the Chromosomally-Encoded mcr-1 Gene. Microorganisms, 7(11), 558. https://doi.org/10.3390/microorganisms7110558