The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution During Fermentation Evaluated by Metagenomic and Culture-Dependent Methods
Abstract
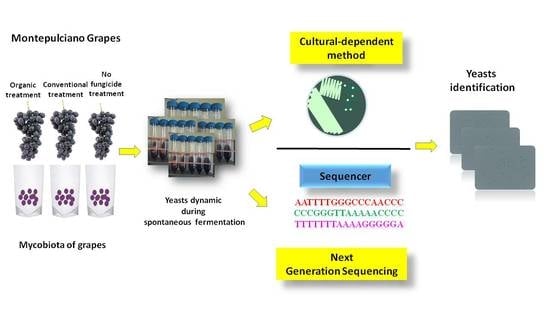
:1. Introduction
2. Materials and Methods
2.1. Vineyard Treatments and Grape Sampling
2.2. Grape Juice Spontaneous Fermentations
2.3. Viable Counts, Yeast Isolations and Analytical Procedures
2.4. Yeasts Identification
2.5. Total DNA Extraction and Next Generation Sequencing (NGS) Analysis
2.6. NGS Data Processing
2.7. Statistical Analysis
3. Results
3.1. Effects of Fungicide Treatments on Fungal Community at Harvest Time
3.1.1. Culture-Independent Analysis (NGS)
3.1.2. Culture-Dependent Analysis
3.2. Effects of Fungicide Treatments on Fungal Community at 7th Day of Spontaneous Fermentation
3.2.1. Culture-Independent Analysis (NGS)
3.2.2. Culture-Dependent Analysis
3.3. Effects of Fungicide Treatments on Fungal Community at 15th Day of Spontaneous Fermentation
3.3.1. Culture-Independent Analysis (NGS)
3.3.2. Culture-Dependent Analysis
3.4. Principal Component Analysis
4. Discussion
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Wisniewski, M.; Nicosia, M.G.L.D.; Cacciola, S.O.; Schena, L. Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS ONE 2016, 11, e0160470. [Google Scholar] [CrossRef] [PubMed]
- Madden, A.A.; Epps, M.J.; Fukami, T.; Irwin, R.E.; Sheppard, J.; Sorger, D.M.; Dunn, R.R. The ecology of insect–yeast relationships and its relevance to human industry. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172733. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [PubMed]
- Comitini, F.; Ciani, M. Influence of fungicide treatments on the occurrence of yeast flora associated with wine grapes. Ann. Microbiol. 2008, 58, 489–493. [Google Scholar] [CrossRef]
- Chavan, P.; Mane, S.; Kulkarni, G.; Shaikh, S.; Ghormade, V.; Nerkar, D.P.; Shouche, Y.; Deshpande, M.V. Natural yeast flora of different varieties of grapes used for wine making in India. Food Microbiol. 2009, 26, 801–808. [Google Scholar] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [PubMed]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [PubMed]
- Zhang, J.; Wang, E.T.; Singh, R.P.; Guo, C.; Shang, Y.; Chen, J.; Liu, C. Grape berry surface bacterial microbiome: impact from the varieties and clones in the same vineyard from central China. J. Appl. Microbiol. 2019, 126, 204–214. [Google Scholar]
- Swiegers, J.H.; Pretorius, I.S. Yeast modulation of wine flavor. Adv. Appl. Microbiol. 2005, 57, 131–175. [Google Scholar]
- Francesca, N.; Canale, D.E.; Settanni, L.; Moschetti, G. Dissemination of wine-related yeasts by migratory birds. Environ. Microbiol. Rep. 2012, 4, 105–112. [Google Scholar] [CrossRef]
- Stefanini, I.; Dapporto, L.; Legras, J.L.; Calabretta, A.; Di Paola, M.; De Filippo, C.; Viola, R.; Capretti, P.; Polsinelli, M.; Tuillazzi, S.; et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-López, F.N.; Salvadó, Z.; Tronchoni, J.; Guillamón, J.M.; Barrio, E.; Querol, A. Susceptibility and resistance to ethanol in Saccharomyces strains isolated from wild and fermentative environments. Yeast 2010, 27, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, V.; Querol, A. The prevalence and control of spoilage yeasts in foods and beverages. Trends Food Sci. Technol. 1999, 10, 356–365. [Google Scholar] [CrossRef]
- Malfeito-Ferreira, M. Yeasts and wine off-flavours: A technological perspective. Ann. Microbiol. 2011, 61, 95–102. [Google Scholar] [CrossRef]
- Regueiro, L.A.; Costas, C.L.; Rubio, J.E.L. Influence of viticultural and enological practices on the development of yeast populations during winemaking. Am. J. Enol. Viticult. 1993, 44, 405–408. [Google Scholar]
- Viviani-Nauer, A.; Hoffman-Boller, P.; Basler, P.; Gafner, J. Wild yeast flora on grapes of fungi disease resistant cultivars and their dynamics during fermentation. Schweiz. Z. Obst-Weinbau 1995, 131, 390–393. [Google Scholar]
- Ganga, M.A.; Martinez, C. Effect of wine yeast monoculture practice on the biodiversity of non-Saccharomyces yeasts. J. Appl. Microbiol. 2004, 96, 76–83. [Google Scholar] [CrossRef]
- Valero, E.; Cambon, B.; Schuller, D.; Casal, M.; Dequin, S. Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Y. Res. 2007, 7, 317–329. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Arroyo, T.; Serrano, A.; Tello, J.; Aporta, I.; Vélez, M.D.; Valero, E. Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 2011, 145, 132–139. [Google Scholar] [CrossRef]
- Tello, J.; Cordero-Bueso, G.; Aporta, I.; Cabellos, J.M.; Arroyo, T. Genetic diversity in commercial wineries: Effects of the farming system and vinification management on wine yeasts. J. Appl. Microbiol. 2012, 112, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Milanović, V.; Comitini, F.; Ciani, M. Grape berry yeast communities: Influence of fungicide treatments. Int J. Food Microbiol. 2013, 161, 240–246. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R.; González-Arenzana, L. Impact of Chemical and Biological Fungicides Applied to Grapevine on Grape Biofilm, Must, and Wine Microbial Diversity. Front. Microbiol. 2018, 9, 59. [Google Scholar] [CrossRef]
- Piao, H.; Hawley, E.; Kopf, S.; De Scenzo, R.; Sealock, S.; Henick-Kling, T.; Hess, M. Insights into the bacterial community and its temporal succession during the fermentation of wine grapes. Front. Microbiol. 2015, 6, 809. [Google Scholar] [CrossRef]
- Cocolin, L.; Alessandria, V.; Dolci, P.; Gorra, R.; Rantsiou, K. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int. J. Food Microbiol. 2013, 167, 29–43. [Google Scholar] [CrossRef]
- Maturano, Y.P.; Mestre, M.V.; Combina, M.; Toro, M.E.; Vazquez, F.; Esteve-Zarzoso, B. Culture-dependent and independent techniques to monitor yeast species during cold soak carried out at different temperatures in winemaking. Int. J. Food Microbiol. 2016, 237, 142–149. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Joseph, C.M.L.; Allen, G.R.; Benson, A.K.; Mills, D.A. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS ONE 2012, 7, e36357. [Google Scholar] [CrossRef]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Exploring the sources of bacterial spoilers in beef steaks by culture-independent high-throughput sequencing. PLoS ONE 2013, 8, e70222. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; García-Fernández, D.; Mas, A.; Esteve-Zarzoso, B. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 2015, 6, 1156. [Google Scholar] [CrossRef] [PubMed]
- Stringini, M.; Comitini, F.; Taccari, M.; Ciani, M. Yeast diversity in crop-growing environments in Cameroon. Int. J. Food Microbiol. 2008, 127, 184–189. [Google Scholar] [CrossRef] [PubMed]
- EEC. Council Regulation 2870/00 laying down Community reference methods for the analysis of spirit drinks. Off. J. Eur. Comm. 2000, L333, 20–46. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8 S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Evol. Microbiol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Envirol. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Verginer, M.; Leitner, E.; Berg, G. Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 2010, 58, 8344–8350. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Rantsiou, K.; Iliopoulos, V.; Cocolin, L.; Nychas, G.J.E. Bacterial species associated with sound and Botrytis-infected grapes from a Greek vineyard. Int. J. Food Microbiol. 2011, 145, 432–436. [Google Scholar] [CrossRef]
- Rantsiou, K.; Urso, R.; Iacumin, L.; Cantoni, C.; Cattaneo, P.; Comi, G.; Cocolin, L. Culture-dependent and-independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 2005, 71, 1977–1986. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the diversity of grapevine microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Valera, M.J.; Torija, M.J.; Mas, A.; Mateo, E. Acetic acid bacteria from biofilm of strawberry vinegar visualized by microscopy and detected by complementing culture-dependent and culture-independent techniques. Food Microbiol. 2015, 46, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Čadež, N.; Zupan, J.; Raspor, P. The effect of fungicides on yeast communities associated with grape berries. FEMS Yeast Res. 2010, 10, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Setati, M.E.; Jacobson, D.; Andong, U.C.; Bauer, F. The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 2013, 7, e52609. [Google Scholar] [CrossRef]
- Ghosh, S.; Bagheri, B.; Morgan, H.H.; Divol, B.; Setati, M.E. Assessment of wine microbial diversity using ARISA and cultivation-based methods. Ann. Microbiol. 2015, 65, 1833–1840. [Google Scholar] [CrossRef]
- Beltran, G.; Torija, M.J.; Novo, M.; Ferrer, N.; Poblet, M.; Guillamón, J.M.; Rozès, N.; Mas, A. Analysis of yeast populations during alcoholic fermentation: A six year follow-up study. Syst. Appl. Microbiol. 2002, 25, 287–293. [Google Scholar] [CrossRef] [PubMed]
- David, V.; Terrat, S.; Herzine, K.; Claisse, O.; Rousseaux, S.; Tourdot-Maréchal, R.; Masneuf-Pomarede, I.; Ranjard, L.; Alexandre, H. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 2014, 41, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Kioroglou, D.; LLeixá, J.; Mas, A.; Portillo, M. Massive sequencing: A new tool for the control of alcoholic fermentation in wine? Fermentation 2018, 4, 7. [Google Scholar] [CrossRef]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Rosini, G.; Federici, F.; Martini, A. Yeast flora of grape berries during ripening. Microbial. Ecol. 1982, 8, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Martini, A.; Ciani, M.; Scorzetti, G. Direct enumeration and isolation of wine yeasts from grape surfaces. Am. J. Enol. Vitic. 1996, 47, 435–440. [Google Scholar]
- Fleet, G.H.; Prakitchaiwattana, C.; Beh, A.L.; Heard, G. The yeast ecology of wine grapes. Biodiversity and biotechnology of wine yeasts. Res. Signpost 2002, 95, 1–17. [Google Scholar]
 ), MC (
), MC (  ) and MNT (
) and MNT (  ) grapes. The results were obtained using the Shannon index. (a–c) represent rarefaction curves and box-and-whisker plots referred to harvest time and at the 7th and 15th day of spontaneous fermentation, respectively.
) grapes. The results were obtained using the Shannon index. (a–c) represent rarefaction curves and box-and-whisker plots referred to harvest time and at the 7th and 15th day of spontaneous fermentation, respectively.
 ), MC (
), MC (  ) and MNT (
) and MNT (  ) grapes. The results were obtained using the Shannon index. (a–c) represent rarefaction curves and box-and-whisker plots referred to harvest time and at the 7th and 15th day of spontaneous fermentation, respectively.
) grapes. The results were obtained using the Shannon index. (a–c) represent rarefaction curves and box-and-whisker plots referred to harvest time and at the 7th and 15th day of spontaneous fermentation, respectively.






 ; conventional (MC)
; conventional (MC)  and not treated (MNT)
and not treated (MNT)  samples at the harvest time (a), at 7th day (b) and at 15th day (c) of spontaneous fermentation. Percentages shown along the axes represent the proportion of dissimilarities captured by the axes.
samples at the harvest time (a), at 7th day (b) and at 15th day (c) of spontaneous fermentation. Percentages shown along the axes represent the proportion of dissimilarities captured by the axes.
 ; conventional (MC)
; conventional (MC)  and not treated (MNT)
and not treated (MNT)  samples at the harvest time (a), at 7th day (b) and at 15th day (c) of spontaneous fermentation. Percentages shown along the axes represent the proportion of dissimilarities captured by the axes.
samples at the harvest time (a), at 7th day (b) and at 15th day (c) of spontaneous fermentation. Percentages shown along the axes represent the proportion of dissimilarities captured by the axes.
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarbati, A.; Canonico, L.; Mancabelli, L.; Milani, C.; Ventura, M.; Ciani, M.; Comitini, F. The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution During Fermentation Evaluated by Metagenomic and Culture-Dependent Methods. Microorganisms 2019, 7, 114. https://doi.org/10.3390/microorganisms7050114
Agarbati A, Canonico L, Mancabelli L, Milani C, Ventura M, Ciani M, Comitini F. The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution During Fermentation Evaluated by Metagenomic and Culture-Dependent Methods. Microorganisms. 2019; 7(5):114. https://doi.org/10.3390/microorganisms7050114
Chicago/Turabian StyleAgarbati, Alice, Laura Canonico, Leonardo Mancabelli, Christian Milani, Marco Ventura, Maurizio Ciani, and Francesca Comitini. 2019. "The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution During Fermentation Evaluated by Metagenomic and Culture-Dependent Methods" Microorganisms 7, no. 5: 114. https://doi.org/10.3390/microorganisms7050114
APA StyleAgarbati, A., Canonico, L., Mancabelli, L., Milani, C., Ventura, M., Ciani, M., & Comitini, F. (2019). The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution During Fermentation Evaluated by Metagenomic and Culture-Dependent Methods. Microorganisms, 7(5), 114. https://doi.org/10.3390/microorganisms7050114