Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Trial
2.2. DNA Extraction and Sequencing
2.3. Bioinformatic Analysis and Statistics
2.4. Accession Number
3. Results
3.1. Effect of In-Feed Antimicrobials on the Cecal Bacterial Diversity
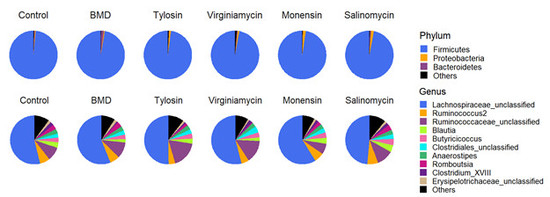
3.2. Effect of In-Feed Antimicrobials on Cecal Bacterial Composition
3.3. Differential Regulation of OTUs by Antimicrobial Supplementation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Seal, B.S.; Lillehoj, H.S.; Donovan, D.M.; Gay, C.G. Alternatives to antibiotics: A symposium on the challenges and solutions for animal production. Anim. Health Res. Rev. 2013, 14, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Broom, L.J. The sub-inhibitory theory for antibiotic growth promoters. Poult. Sci. 2017, 96, 3104–3108. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.D.; Butcher, G.D.; Henry, P.R.; Littell, R.C. Effect of antibiotic growth promoters on broiler performance, intestinal growth parameters, and quantitative morphology. Poult. Sci. 2006, 85, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Lhermie, G.; Grohn, Y.T.; Raboisson, D. Addressing antimicrobial resistance: An overview of priority actions to prevent suboptimal antimicrobial use in food-animal production. Front. Microbiol. 2016, 7, 2114. [Google Scholar] [CrossRef] [PubMed]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. Mbio 2016, 7, e02227-15. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Stanton, T.B. Altered egos: Antibiotic effects on food animal microbiomes. Annu. Rev. Microbiol. 2014, 68, 297–315. [Google Scholar] [CrossRef]
- Yadav, S.; Jha, R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J. Anim. Sci. Biotechnol. 2019, 10, 2. [Google Scholar] [CrossRef]
- Angelakis, E. Weight gain by gut microbiota manipulation in productive animals. Microb. Pathog. 2017, 106, 162–170. [Google Scholar] [CrossRef]
- Coates, M.E.; Fuller, R.; Harrison, G.F.; Lev, M.; Suffolk, S.F. A comparison of the growth of chicks in the gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br. J. Nutr. 1963, 17, 141–150. [Google Scholar] [CrossRef]
- Lalles, J.P. Microbiota-host interplay at the gut epithelial level, health and nutrition. J. Anim. Sci. Biotechnol. 2016, 7, 66. [Google Scholar] [CrossRef]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, S.; Wang, Y.; Cai, S.; Yu, H.; Liu, H.; Zeng, X.; Zhang, G.; Qiao, S. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol. Life Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra-Hijar, G.; Abo-Ismail, M.; Carstens, G.E.; Guan, L.L.; Hegarty, R.; Kenny, D.A.; McGee, M.; Plastow, G.; Relling, A.; Ortigues-Marty, I. Review: Biological determinants of between-animal variation in feed efficiency of growing beef cattle. Animal 2018, 12, s321–s335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Munoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef] [PubMed]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.J.; Billon, Y.; Berri, M.; Dore, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.G.; Kim, E.B.; Lee, J.; Lee, J.Y.; Jin, G.; Park, J.; Huh, C.S.; Kwon, I.K.; Kil, D.Y.; Choi, Y.J. Relationship between the microbiota in different sections of the gastrointestinal tract, and the body weight of broiler chickens. Springerplus 2016, 5, 911. [Google Scholar] [CrossRef] [PubMed]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, R Version 3.5.1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.5-2. 2018. Available online: https://github.com/vegandevs/vegan (accessed on 17 May 2018).
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. Venndiagram: A package for the generation of highly-customizable venn and euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Dumonceaux, T.J.; Hill, J.E.; Hemmingsen, S.M.; Van Kessel, A.G. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 2006, 72, 2815–2823. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, A.A.; Menten, J.F.; Lambais, M.R.; Racanicci, A.M.; Longo, F.A.; Sorbara, J.O. Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult. Sci. 2006, 85, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gong, J.; Brisbin, J.T.; Yu, H.; Sanei, B.; Sabour, P.; Sharif, S. Appropriate chicken sample size for identifying the composition of broiler intestinal microbiota affected by dietary antibiotics, using the polymerase chain reaction-denaturing gradient gel electrophoresis technique. Poult. Sci. 2007, 86, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Hofacre, C.; Smith, F.; Lee, M.D. Effects of feed additives on the development on the ileal bacterial community of the broiler chicken. Animal 2008, 2, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Czerwinski, J.; Hojberg, O.; Smulikowska, S.; Engberg, R.M.; Mieczkowska, A. Effects of sodium butyrate and salinomycin upon intestinal microbiota, mucosal morphology and performance of broiler chickens. Arch. Anim. Nutr. 2012, 66, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Hunkapiller, A.A.; Layton, A.C.; Chang, Y.J.; Robbins, K.R. Response of intestinal microbiota to antibiotic growth promoters in chickens. Foodborne Pathog. Dis. 2013, 10, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Danzeisen, J.L.; Kim, H.B.; Isaacson, R.E.; Tu, Z.J.; Johnson, T.J. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE 2011, 6, e27949. [Google Scholar] [CrossRef]
- Neumann, A.P.; Suen, G. Differences in major bacterial populations in the intestines of mature broilers after feeding virginiamycin or bacitracin methylene disalicylate. J. Appl. Microbiol. 2015, 119, 1515–1526. [Google Scholar] [CrossRef]
- Costa, M.C.; Bessegatto, J.A.; Alfieri, A.A.; Weese, J.S.; Filho, J.A.; Oba, A. Different antibiotic growth promoters induce specific changes in the cecal microbiota membership of broiler chicken. PLoS ONE 2017, 12, e0171642. [Google Scholar] [CrossRef] [PubMed]
- Proctor, A.; Phillips, G.J. Differential effects of bacitracin methylene disalicylate (bmd) on the distal colon and cecal microbiota of young broiler chickens. Front. Vet. Sci. 2019, 6, 114. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, K.; Kim, D.W.; Kil, D.Y.; Kim, G.B.; Cha, C.J. Influence of dietary avilamycin on ileal and cecal microbiota in broiler chickens. Poult. Sci. 2018, 97, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Diaz Carrasco, J.M.; Redondo, E.A.; Pin Viso, N.D.; Redondo, L.M.; Farber, M.D.; Fernandez Miyakawa, M.E. Tannins and bacitracin differentially modulate gut microbiota of broiler chickens. BioMed Res. Int 2018, 2018, 1879168. [Google Scholar] [CrossRef]
- Crisol-Martinez, E.; Stanley, D.; Geier, M.S.; Hughes, R.J.; Moore, R.J. Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: Linking gut microbiota and growth performance in chickens. Appl. Microbiol. Biotechnol. 2017, 101, 4547–4559. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, C.I.; Woodward, M.J.; Ellis, R.J.; La Ragione, R.M.; Claus, S.P. Antibiotic treatment triggers gut dysbiosis and modulates metabolism in a chicken model of gastro-intestinal infection. BMC Vet. Res. 2019, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Pourabedin, M.; Guan, L.; Zhao, X. Xylo-oligosaccharides and virginiamycin differentially modulate gut microbial composition in chickens. Microbiome 2015, 3, 15. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Banerjee, S.; Sar, A.; Misra, A.; Pal, S.; Chakraborty, A.; Dam, B. Increased productivity in poultry birds by sub-lethal dose of antibiotics is arbitrated by selective enrichment of gut microbiota, particularly short-chain fatty acid producers. Microbiology 2018, 164, 142–153. [Google Scholar] [CrossRef]
- Torok, V.A.; Allison, G.E.; Percy, N.J.; Ophel-Keller, K.; Hughes, R.J. Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance. Appl. Environ. Microbiol. 2011, 77, 3380–3390. [Google Scholar] [CrossRef]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: Acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Ljungh, A.; Wadstrom, T. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 2006, 7, 73–89. [Google Scholar] [PubMed]
- Wan, L.Y.; Chen, Z.J.; Shah, N.P.; El-Nezami, H. Modulation of intestinal epithelial defense responses by probiotic bacteria. Crit. Rev. Food Sci. Nutr. 2016, 56, 2628–2641. [Google Scholar] [CrossRef] [PubMed]
- Guban, J.; Korver, D.R.; Allison, G.E.; Tannock, G.W. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult. Sci. 2006, 85, 2186–2194. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. Antibiotic growth promoters enhance animal production by targeting intestinal bile salt hydrolase and its producers. Front. Microbiol. 2014, 5, 33. [Google Scholar] [CrossRef]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef]
- Allen, H.K.; Looft, T.; Bayles, D.O.; Humphrey, S.; Levine, U.Y.; Alt, D.; Stanton, T.B. Antibiotics in feed induce prophages in swine fecal microbiomes. Mbio 2011, 2, e00260-11. [Google Scholar] [CrossRef]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Antoszczak, M.; Steverding, D.; Huczynski, A. Anti-parasitic activity of polyether ionophores. Eur. J. Med. Chem. 2019, 166, 32–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Sheikhly, F.; Al-Saieg, A. Role of coccidia in the occurrence of necrotic enteritis of chickens. Avian Dis. 1980, 24, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Collier, C.T.; Hofacre, C.L.; Payne, A.M.; Anderson, D.B.; Kaiser, P.; Mackie, R.I.; Gaskins, H.R. Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet. Immunol. Immunopathol. 2008, 122, 104–115. [Google Scholar] [CrossRef] [PubMed]






| Control | BMD | Tylosin | Virginiamycin | Monensin | Salinomycyin | |
|---|---|---|---|---|---|---|
| Control | p = 0.043 R = 0.071 | p = 0.052 R = 0.061 | p = 0.036 R = 0.090 | p < 0.001 R = 0.921 | p < 0.001 R = 0.809 | |
| BMD | p = 0.013 1 R = 0.145 | p = 0.243 R = 0.021 | p = 0.043 R = 0.095 | p < 0.001 R = 0.914 | p < 0.001 R = 0.774 | |
| Tylosin | p = 0.009 R = 0.149 | p = 0.004 R = 0.187 | p = 0.595 R = 0.015 | p < 0.001 R = 0.855 | p < 0.001 R = 0.714 | |
| Virginiamycin | p = 0.001 R = 0.226 | p = 0.009 R = 0.164 | p = 0.197 R = 0.048 | p < 0.001 R = 0.957 | p < 0.001 R = 0.832 | |
| Monensin | p < 0.001 R = 0.315 | p < 0.001 R = 0.284 | p < 0.001 R = 0.411 | p < 0.001 R = 0.401 | p = 0.021 R = 0.076 | |
| Salinomycin | p = 0.002 R = 0.163 | p = 0.003 R = 0.205 | p < 0.001 R = 0.294 | p < 0.001 R = 0.292 | p = 0.029 R = 0.105 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, K.; Becker, S.; Xiao, Y.; Lyu, W.; Yang, Q.; Zhu, H.; Yang, H.; Zhao, J.; Zhang, G. Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers. Microorganisms 2019, 7, 282. https://doi.org/10.3390/microorganisms7090282
Robinson K, Becker S, Xiao Y, Lyu W, Yang Q, Zhu H, Yang H, Zhao J, Zhang G. Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers. Microorganisms. 2019; 7(9):282. https://doi.org/10.3390/microorganisms7090282
Chicago/Turabian StyleRobinson, Kelsy, Sage Becker, Yingping Xiao, Wentao Lyu, Qing Yang, Huiling Zhu, Hua Yang, Jiangchao Zhao, and Guolong Zhang. 2019. "Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers" Microorganisms 7, no. 9: 282. https://doi.org/10.3390/microorganisms7090282
APA StyleRobinson, K., Becker, S., Xiao, Y., Lyu, W., Yang, Q., Zhu, H., Yang, H., Zhao, J., & Zhang, G. (2019). Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers. Microorganisms, 7(9), 282. https://doi.org/10.3390/microorganisms7090282