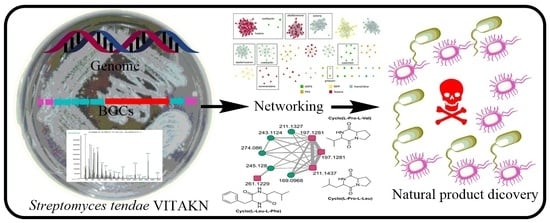
Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Actinomycetes
2.2. Morphological and Cultural Characteristics of the Strain
2.3. Crude Extracts Preparation and Quorum Sensing Inhibitory (QSI) Activity
2.4. Genomic Characterization
2.5. HPLC-HRMS/MS Analysis and Metabolites Identification Using GNPS Dereplication
2.6. Data Deposition and Job Accessibility
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benndorf, R.; Guo, H.; Sommerwerk, E.; Weigel, C.; Garcia-Altares, M.; Martin, K.; Hu, H.; Kufner, M.; de Beer, Z.W.; Poulsen, M.; et al. Natural Products from Actinobacteria Associated with Fungus-Growing Termites. Antibiotics 2018, 7, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [Green Version]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdorf, I.; Oliviero, G.; Costantino, V.; Morgenstern, D.; Steindler, L. In Search of Alternative Antibiotic Drugs: Quorum-Quenching Activity in Sponges and their Bacterial Isolates. Front. Microbiol. 2016, 7, 416. [Google Scholar] [CrossRef] [Green Version]
- Della Sala, G.; Teta, R.; Esposito, G.; Costantino, V. Chapter 1—The Chemical Language of Gram-Negative Bacteria. In Quorum Sensing; Tommonaro, G., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 3–28. [Google Scholar]
- Strömstedt, A.A.; Felth, J.; Bohlin, L. Bioassays in Natural Product Research—Strategies and Methods in the Search for Anti-inflammatory and Antimicrobial Activity. Phytochem. Anal. 2014, 25, 13–28. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Mikheenko, A.; Garg, N.; Nothias, L.F.; Ninomiya, A.; Takada, K.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017, 13, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [Green Version]
- Schorn, M.A.; Alanjary, M.M.; Aguinaldo, K.; Korobeynikov, A.; Podell, S.; Patin, N.; Lincecum, T.; Jensen, P.R.; Ziemert, N.; Moore, B.S. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 2016, 162, 2075–2086. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, G.; Bourguet-Kondracki, M.L.; Mai, L.H.; Longeon, A.; Teta, R.; Meijer, L.; Van Soest, R.; Mangoni, A.; Costantino, V. Chloromethylhalicyclamine B, a Marine-Derived Protein Kinase CK1delta/epsilon Inhibitor. J. Nat. Prod. 2016, 79, 2953–2960. [Google Scholar] [CrossRef] [PubMed]
- Gebreyohannes, G.; Moges, F.; Sahile, S.; Raja, N. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac. J. Trop. Biomed. 2013, 3, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Hong, K.; Gao, A.H.; Xie, Q.Y.; Gao, H.; Zhuang, L.; Lin, H.P.; Yu, H.P.; Li, J.; Yao, X.S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef]
- Kumar, S.; Kannabiran, K. Antifungal activity of Streptomyces VITSVK5 spp. against drug resistant Aspergillus clinical isolates from pulmonary tuberculosis patients. J. Mycol. Med. 2010, 20, 101–107. [Google Scholar] [CrossRef]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jorgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192. [Google Scholar] [CrossRef]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum Sensing Inhibitors from the Sea Discovered Using Bacterial N-acyl-homoserine Lactone-Based Biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef] [Green Version]
- Costantino, V.; Della Sala, G.; Saurav, K.; Teta, R.; Bar-Shalom, R.; Mangoni, A.; Steindler, L. Plakofuranolactone as a Quorum Quenching Agent from the Indonesian Sponge Plakortis cf. lita. Mar. Drugs 2017, 15, 59. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res. 2018, 7. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019, 8, giz100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoSComput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based Molecular Networking in the GNPS Analysis Environment. bioRxiv 2019, 812404. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Caso, A.; Esposito, G.; Della Sala, G.; Pawlik, J.R.; Teta, R.; Mangoni, A.; Costantino, V. Fast Detection of Two Smenamide Family Members Using Molecular Networking. Mar. Drugs 2019, 17, 618. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species1. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar]
- Kodani, S.; Hudson, M.E.; Durrant, M.C.; Buttner, M.J.; Nodwell, J.R.; Willey, J.M. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 2004, 101, 11448–11453. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Moody, S.C.; Hider, R.C.; Lei, L.; Kelly, S.L.; Waterman, M.R.; Lamb, D.C. Structural analysis of cytochrome P450 105N1 involved in the biosynthesis of the zincophore, coelibactin. Int. J. Mol. Sci. 2012, 13, 8500–8513. [Google Scholar] [CrossRef] [Green Version]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Omura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2019, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.M.; Bandeira, N.; Shen, Z.; Tanner, S.; Briggs, S.P.; Smith, R.D.; Pevzner, P.A. Clustering millions of tandem mass spectra. J. Proteome Res. 2008, 7, 113–122. [Google Scholar] [CrossRef]
- Campbell, J.; Lin, Q.; Geske, G.D.; Blackwell, H.E. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 2009, 4, 1051–1059. [Google Scholar] [CrossRef]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Gu, Q.; Li, W. Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar] [CrossRef]
- Canu, N.; Moutiez, M.; Belin, P.; Gondry, M. Cyclodipeptide synthases: A promising biotechnological tool for the synthesis of diverse 2,5-diketopiperazines. Nat. Prod. Rep. 2019. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishaque, N.M.; Burgsdorf, I.; Limlingan Malit, J.J.; Saha, S.; Teta, R.; Ewe, D.; Kannabiran, K.; Hrouzek, P.; Steindler, L.; Costantino, V.; et al. Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India. Microorganisms 2020, 8, 121. https://doi.org/10.3390/microorganisms8010121
Ishaque NM, Burgsdorf I, Limlingan Malit JJ, Saha S, Teta R, Ewe D, Kannabiran K, Hrouzek P, Steindler L, Costantino V, et al. Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India. Microorganisms. 2020; 8(1):121. https://doi.org/10.3390/microorganisms8010121
Chicago/Turabian StyleIshaque, Nabila Mohammed, Ilia Burgsdorf, Jessie James Limlingan Malit, Subhasish Saha, Roberta Teta, Daniela Ewe, Krishnan Kannabiran, Pavel Hrouzek, Laura Steindler, Valeria Costantino, and et al. 2020. "Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India" Microorganisms 8, no. 1: 121. https://doi.org/10.3390/microorganisms8010121
APA StyleIshaque, N. M., Burgsdorf, I., Limlingan Malit, J. J., Saha, S., Teta, R., Ewe, D., Kannabiran, K., Hrouzek, P., Steindler, L., Costantino, V., & Saurav, K. (2020). Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India. Microorganisms, 8(1), 121. https://doi.org/10.3390/microorganisms8010121