Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., Isolated from the Discovery Deep Brine-Seawater Interface in the Red Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Growth of Strains SB29T and SB3T
2.2. Selection, Morphology and Phylogeny of Strains
2.3. Phenotypic Tests
2.4. Genomic Analyses
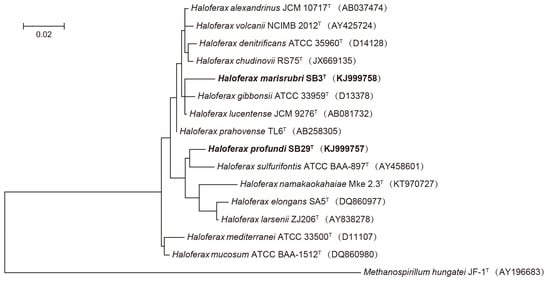
2.5. Phylogenetic Analyses
2.6. Data Availability
3. Results
4. Discussion
4.1. Description of Haloferax profundi sp. nov.
4.2. Description of Haloferax marisrubri sp. nov.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bougouffa, S.; Yang, J.K.; Lee, O.O.; Wang, Y.; Batang, Z.; Al-Suwailem, A.; Qian, P.Y. Distinctive Microbial Community Structure in Highly Stratified Deep-Sea Brine Water Columns. Appl. Environ. Microbiol. 2013, 79, 3425–3437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eder, W.; Jahnke, L.L.; Schmidt, M.; Huber, R. Microbial Diversity of the Brine-Seawater Interface of the Kebrit Deep, Red Sea, Studied via 16S rRNA Gene Sequences and Cultivation Methods. Appl. Environ. Microbiol. 2001, 67, 3077–3085. [Google Scholar] [CrossRef] [Green Version]
- Daffonchio, D.; Borin, S.; Brusa, T.; Brusetti, L.; van der Wielen, P.W.J.J.; Bolhuis, H.; Yakimov, M.M.; D’Auria, G.; Giuliano, L.; Marty, D.; et al. Stratified prokaryote network in the oxic–anoxic transition of a deep-sea halocline. Nature 2006, 440, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Naushad, S.; Baker, S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: A proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1050–1069. [Google Scholar] [PubMed]
- Mullakhanbhai, M.F.; Larsen, H. Halobacteriumvolcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 1975, 104, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Valera, F.; Juez, G.; Kushner, D.J. Halobacterium mediterranei spec, nov., a New Carbohydrate-Utilizing Extreme Halophile. Syst. Appl. Microbiol. 1983, 4, 369–381. [Google Scholar] [CrossRef]
- Ga, T.; Li, J.; Li, H. Halobacterium denitrificans sp. nov., an extremely halophilic denitrifying bacterium. Int. J. Syst. Bacteriol. 1986, 36, 66–70. [Google Scholar]
- Juez, G.; Rodriguez-Valera, F.; Ventosa, A.; Kushner, D.J. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst. Appl. Microbiol. 1986, 8, 75–79. [Google Scholar] [CrossRef]
- Asker, D.; Ohta, Y. Haloferax alexandrinus sp. nov., an extremely halophilic canthaxanthin-producing archaeon from a solar saltern in Alexandria (Egypt). Int. J. Syst. Evol. Microbiol. 2002, 52, 729–738. [Google Scholar]
- Gutierrez, M.C.; Kamekura, M.; Holmes, M.L.; Dyall-Smith, M.L.; Ventosa, A. Taxonomic characterization of Haloferax sp. (“H. alicantei”) strain Aa 2.2: Description of Haloferax lucentensis sp. nov. Extremophiles 2002, 6, 479–483. [Google Scholar] [CrossRef]
- Elshahed, M.S.; Savage, K.N.; Oren, A.; Gutierrez, M.C.; Ventosa, A.; Krumholz, L.R. Haloferax sulfurifontis sp. nov., a halophilic archaeon isolated from a sulfide- and sulfur-rich spring. Int. J. Syst. Evol. Microbiol. 2004, 54, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Enache, M.; Itoh, T.; Kamekura, M.; Teodosiu, G.; Dumitru, L. Haloferax prahovense sp. nov., an extremely halophilic archaeon isolated from a Romanian salt lake. Int. J. Syst. Evol. Microbiol. 2007, 57, 393–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.W.; Wu, Y.H.; Wang, C.S.; Oren, A.; Zhou, P.J.; Wu, M. Haloferax larsenii sp. nov., an extremely halophilic archaeon from a solar saltern. Int. J. Syst. Evol. Microbiol. 2007, 57, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.A.; Goh, F.; Leuko, S.; Echigo, A.; Mizuki, T.; Usami, R.; Kamekura, M.; Neilan, A.B.; Burns, B.P. Haloferax elongans sp. nov. and Haloferax mucosum sp. nov., isolated from microbial mats from Hamelin Pool, Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2008, 58, 798–802. [Google Scholar] [CrossRef] [Green Version]
- Saralov, A.I.; Baslerov, R.V.; Kuznetsov, B.B. Haloferax chudinovii sp. nov., a halophilic archaeon from Permian potassium salt deposits. Extremophiles 2013, 17, 499–504. [Google Scholar] [CrossRef]
- Mcduff, S.; King, G.M.; Neupane, S.; Myers, M.R. Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinders, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3T, sp. nov. FEMS Microbiol. Ecol. 2016, 92, fiw028. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Gu, J.; Zhang, R.; Rashid, M.; Haroon, M.F.; Xun, W.; Ruan, Z.; Dong, X.; Stingl, U. Haloprofundus marisrubri gen. nov., sp. nov., an extremely halophilic archaeon isolated from a brine-seawater interface. Int. J. Syst. Evol. Microbiol. 2017, 67, 9–16. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Oren, A.; Ventosa, A.; Grant, W.D. Proposed minimal standards for description of new taxa in the order Halobacteriales. Int. J. Syst. Bacteriol. 1997, 47, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Rieger, G.; Müller, K.; Hermann, R.; Stetter, K.O.; Rachel, R. Cultivation of hyperthermophilic archaea in capillary tubes resulting in improved preservation of fine structures. Arch. Microbiol. 1997, 168, 373–379. [Google Scholar] [CrossRef]
- Huber, H.; Burggraf, S.; Mayer, T.; Wyschkony, I.; Rachel, R.; Stetter, K.O. Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov and Ignicoccus pacificus sp. nov and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50. [Google Scholar] [CrossRef] [PubMed]
- Dussault, H.P. An improved technique for staining red halophilic bacteria. J. Bacteriol. 1955, 70, 484–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, F.; Leuko, S.; Allen, M.A.; Bowman, J.P.; Kamekura, M.; Neilan, B.A.; Burns, B.P. Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2006, 56, 1323–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, C.; González, C. Method for Simultaneous Detection of Proteinase and Esterase Activities in Extremely Halophilic Bacteria. Appl. Microbiol. 1972, 24, 516–517. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.L.; Gao, X.; Sun, F.F.; Dong, Y.; Xu, X.W.; Zhou, Y.-G.; Liu, H.-C.; Oren, O.; Zhou, P.-J. Halogranum rubrum gen. nov., sp. nov., a halophilic archaeon isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2010, 60, 1366–1371. [Google Scholar] [CrossRef] [Green Version]
- Dyall-Smith, M.L. The Halohandbook: Protocols for Haloarchaeal Genetics. 2008. Available online: http://www.haloarchaea.com/resources/halohandbook/ (accessed on 7 July 2020).
- Du, Z.J.; Wang, Y.; Dunlap, C.; Rooney, A.P.; Chen, G.J. Draconibacterium orientale gen. nov., sp. nov., isolated from two distinct marine environments, and proposal of Draconibacteriaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1690–1696. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.L.; Pyzyna, B.; Atrasz, R.G.; Henderson, C.A.; Morrill, K.L.; Anna Mae, B.; Desoucy, E.; Fogleman, R.E., 3rd; Naylor, J.B.; Steele, S.M.; et al. Growth kinetics of extremely halophilic Archaea (family Halobacteriaceae) as revealed by Arrhenius plots. J. Bacteriol. 2005, 187, 923–929. [Google Scholar] [CrossRef] [Green Version]
- Stan-Lotter, H.; Pfaffenhuemer, M.; Legat, A.; Busse, H.J.; Radax, C.; Gruber, C. Halococcus dombrowskii sp. nov., an archaeal isolate from a Permian alpine salt deposit. Int. J. Syst. Evol. Microbiol. 2002, 52, 1807–1814. [Google Scholar]
- Jackman, P.J.H. Microbial systematics based on electrophoretic whole-cell protein patterns. Methods Microbiol. 1987, 19, 209–225. [Google Scholar]
- Bligh, E.G. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Tindall, B.J.; Sikorski, J.; Smibert, R.M.; Krieg, N.R. Phenotypic characterization and the principles of comparative systematics. In Methods for General and Molecular Microbiology, 3rd ed.; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G., Marzluf, T.M., Schmidt, L.R., Eds.; Snyder ASM Press: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Liu, N.; Li, Z.; Chen, Y.; Mu, D.; Fan, W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 2013, arXiv:1308.2012. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.; Morre, L.H.; Morre, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef] [Green Version]
- Graham, P.H.; Sadowsky, M.J.; Keyser, H.H.; Barnet, Y.M.; Bradley, R.S.; Cooper, J.E.; de Ley, D.J.; Jarvis, B.D.W.; Roslycky, E.B.; Strijdom, B.W.; et al. Proposed minimal standards for the description of new genera and species of root- and stem-nodulating bacteria. Int. J. Syst. Bacteriol. 1991, 41, 582–587. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. SpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Minegishi, H.; Kamekura, M.; Kitajima-Ihara, T.; Nakasone, K.; Echigo, A.; Shimane, Y.; Usami, R.; Itoh, T.; Ihara, K. Gene orders in the upstream of 16S rRNA genes divide genera of the family Halobacteriaceae into two groups. Int. J. Syst. Evol. Microbiol. 2012, 62, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamekura, M.; Mizuki, T.; Usami, R.; Yoshida, Y.; Horikoshi, K.; Vreeland, V.H. The potential use of signature bases from 16S rRNA gene sequences to aid the assignment of microbial strains to genera of halobacteria. In Halophilic Microorganisms; Ventosa, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 77–100. [Google Scholar]

| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pigmentation | Pink | Pink-Red | Red | Pink-Red | Pink | Red-Orange | Pink | Orange-Red | Orange-Red | Red | Salmon-Pink | Beige-Orange | Orange-Red | Pink-Red | Red |
| Motility | + | + | Rotating | − | + | Rotating | + | − | + | − | − | − | + | − | − |
| NaCl range (M) | 1.0–5.5 | 1.5–5.8 | 1.7–5.1 | 1.7–5.1 | 1.3–4.7 | 1.0–4.5 | 1.8–5.1 | 1.5–4.5 | 1.5–5.2 | 1.7–5.2 | 1.0–5.2 | 2.5–5.2 | 1.0–4.8 | 1.1–4.6 | 0.5–5.4 |
| NaCl optimum (M) | 3.5–4.5 | 4.5–5.0 | 2.6–3.4 | 2.6–3.4 | 2.9 | 1.7–2.5 | 4.3 | 2.0–3.0 | 2.5–4.3 | 4.3 | 2.1–2.6 | 3.5 | 2.2–3.4 | 2.5–3.0 | 1–2 |
| Minimum Mg2+ (M) | 0.2 | 0.35 | 0.2 | 0.2 | 0.02 | 0.02 | 0.07 | 0.06 | 0.2 | 0.33 | 0.001 | 0.1 | 0.005 | 0.02 | nd |
| Temp. range (°C) | 15–45 | 20–50 | 30–55 | 23–55 | 25–45 | 20–45 | 10–40 | 30–55 | 25–55 | 20–55 | 18–50 | 23–51 | 25–55 | 23–51 | nd |
| Temp. optimum (°C) | 33 | 37 | 53 | 42–53 | 35–37 | 45 | 37 | 50 | 35–40 | 37 | 32–37 | 38–48 | 42–45 | 40–45 | 30 |
| pH range | 5.5–9.0 | 6.5–9.0 | 7.0–9.0 | 6.0–10.0 | 5.5–8.0 | 6.0–8.0 | 5.0–9.0 | 6.0–8.0 | 5.0–8.0 | 5.5–7.5 | 5.0–9.0 | 6.0–8.5 | 6.0–8.5 | 5.5–8.0 | nd # |
| Oxidase test | + | + | ± | − | + | + | + | + | + | + | + | + | + | ± | + |
| H2S formation | |||||||||||||||
| from thiosulfate | − | − | − | − | − | + | + | + | + | + | + | + | + | − | nd |
| Hydrolysis of: | |||||||||||||||
| Gelatin | + | + | + | + | + | − | − | + | + | + | + | − | + | − | nd |
| Casein | + | − | + | + | + | − | − | − | + | − | − | − | − | − | nd |
| Starch | + | + | + | − | + | − | − | − | − | − | − | + | + | + | − |
| Tween 80 | − | − | + | − | + | − | + | − | + | + | + | + | + | + | nd |
| Acid production from: | |||||||||||||||
| D-glucose | − | − | − | − | − | + | − | − | + | + | − | + | − | − | nd |
| Mannose | − | − | − | − | + | − | − | − | + | − | − | + | − | − | nd |
| Galactose | − | − | − | − | + | + | − | + | + | − | + | − | − | − | nd |
| Xylose | − | − | − | − | + | + | + | − | + | + | + | − | − | + | nd |
| Sucrose | + | − | + | + | + | + | − | + | + | + | + | − | w | − | nd |
| DNA G + C content | |||||||||||||||
| (mol%) | 60.75 | 65.64 | 61.2 | 61.8 | 60.25 | 65.63 | 66.4 | 66.3 | 66.07 | 66.15 | 66.3 | 65.7 | 61.8 | 65.0 * | 61.5 * |
| DNA-DNA reassociation (DDH): | |||||||||||||||
| with SB29T (%) | 100 | 32.1 | 41.8 | 38.2 | 31.2 | 37.4 | 43.2 | 35.5 | 42.1 | 32.8 | 41.7 | 34.8 | 38.9 | 31.6 | 30.9 |
| with SB3T (%) | 32.1 | 100 | 36.8 | 45.2 | 32.9 | 43.5 | 44.7 | 43.9 | 37.8 | 44.1 | 42.3 | 38.3 | 34.7 | 46.3 | 35.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Dong, X.; Sun, Y.; Antunes, A.; Hikmawan, T.; Haroon, M.F.; Wang, J.; Stingl, U. Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., Isolated from the Discovery Deep Brine-Seawater Interface in the Red Sea. Microorganisms 2020, 8, 1475. https://doi.org/10.3390/microorganisms8101475
Zhang G, Dong X, Sun Y, Antunes A, Hikmawan T, Haroon MF, Wang J, Stingl U. Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., Isolated from the Discovery Deep Brine-Seawater Interface in the Red Sea. Microorganisms. 2020; 8(10):1475. https://doi.org/10.3390/microorganisms8101475
Chicago/Turabian StyleZhang, Guishan, Xiaoyan Dong, Yingjiao Sun, André Antunes, Tyas Hikmawan, Mohamed Fauzi Haroon, Junru Wang, and Ulrich Stingl. 2020. "Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., Isolated from the Discovery Deep Brine-Seawater Interface in the Red Sea" Microorganisms 8, no. 10: 1475. https://doi.org/10.3390/microorganisms8101475
APA StyleZhang, G., Dong, X., Sun, Y., Antunes, A., Hikmawan, T., Haroon, M. F., Wang, J., & Stingl, U. (2020). Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., Isolated from the Discovery Deep Brine-Seawater Interface in the Red Sea. Microorganisms, 8(10), 1475. https://doi.org/10.3390/microorganisms8101475