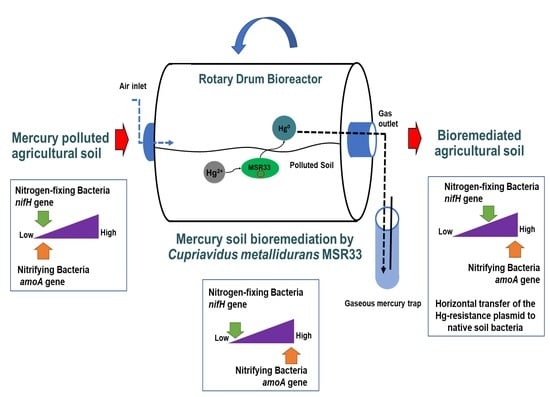
Bioremediation by Cupriavidus metallidurans Strain MSR33 of Mercury-Polluted Agricultural Soil in a Rotary Drum Bioreactor and Its Effects on Nitrogen Cycle Microorganisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Strains
2.3. Agricultural Soil Samples
2.4. Preparation of Mercury-Polluted Agricultural Soil
2.5. Batch Culture Growth
2.6. Bioremediation of Mercury-Polluted Agricultural Soils in a Rotary Drum Bioreactor (RDB)
2.7. Soil Mercury Determination
2.8. Bacterial Count and Isolation of Mercury-Tolerant Strains
2.9. Genomic DNA Extraction from Bacterial Isolates and PCR Amplification of 16S rRNA and merG Genes
2.10. 16S rRNA Gene Sequence Analyses
2.11. Metagenomic DNA Extraction from Agricultural Soil
2.12. Quantification of Nitrogen Cycle-Associated Bacteria and Strain MSR33 in Agricultural Soils
2.13. Statistical Analysis
3. Results
3.1. Evaluation of Operational Parameters on Bioremediation by Strain MSR33 of Mercury-Polluted Agricultural Soil in an RDB
3.2. Effects of Mercury and Mercury Bioremediation on Bacterial Communities of Agricultural Soils
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abhilash, P.C.; Kant Dubey, R.; Tripathi, V.; Gupta, V.K.; Singh, H.B. Plant growth promoting microorganisms for environmental sustainability. Trends Biotechnol. 2016, 34, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Jia, Z.; Conrad, R.; Seeger, M. Simazine application inhibits nitrification and changes the ammonia-oxidizing bacterial communities in a fertilized agricultural soil. FEMS Microbiol. Ecol. 2011, 78, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altimira, F.; Yáñez, C.; Bravo, G.; González, M.; Rojas, L.A.; Seeger, M. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of Central Chile. BMC Microbiol. 2012, 12, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seshadri, B.; Bolan, N.; Naidu, R. Rhizosphere-induced heavy metal(loid) transformation in relation to bioavailability and remediation. J. Soil Sci. Plant Nutr. 2015, 15, 524–548. [Google Scholar] [CrossRef] [Green Version]
- Tosi, M.; Brown, S.; Ferrari Machado, P.; Wagner-Riddle, C.; Dunfield, K. Short-term response of soil N-cycling genes and transcripts to fertilization with nitrification and urease inhibitors, and relationship with field-scale N2O emissions. Soil Biol. Biochem. 2020, 142, 107703. [Google Scholar] [CrossRef]
- Hai, B.; Diallo, N.H.; Sall, S.; Haesler, F.; Schauss, K.; Bonzi, M.; Assigbetse, K.; Chotte, J.-L.; Munch, J.C.; Schloter, M. Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl. Environ. Microbiol. 2009, 75, 4993–5000. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition—Current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [Green Version]
- Rütting, T.; Aronsson, H.; Delin, S. Efficient use of nitrogen in agriculture. Nutr. Cycl. Agroecosyst. 2018, 110, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Filip, Z. International approach to assessing soil quality by ecologically-related biological parameters. Agric. Ecosyst. Environ. 2002, 88, 169–174. [Google Scholar] [CrossRef]
- Orlando, J.; Alfaro, M.; Bravo, L.; Guevara, R.; Carú, M. Bacterial diversity and occurrence of ammonia-oxidizing bacteria in the Atacama desert soil during a ‘desert bloom’ event. Soil Biol. Biochem. 2010, 42, 1183–1188. [Google Scholar] [CrossRef]
- Menéndez, E.; Paço, A. Is the application of plant probiotic bacterial consortia always beneficial for plants? exploring synergies between rhizobial and non-rhizobial bacteria and their effects on agro-economically valuable crops. Life 2020, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Yannarell, A.C.; Mackie, R.I. Changes in N-transforming Archaea and Bacteria in soil during the establishment of bioenergy crops. PLoS ONE 2011, 6, e24750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salam, L.N.; Shomope, H.; Ummi, Z.; Bukar, F. Mercury contamination imposes structural shift on the microbial community of an agricultural soil. Bull. Natl. Res. Cent. 2019, 43, 163. [Google Scholar] [CrossRef]
- Patra, M.; Sharma, A. Mercury toxicity in plants. Bot. Rev. 2000, 66, 379–422. [Google Scholar] [CrossRef]
- Streets, D.G.; Horowitz, H.M.; Jacob, D.J.; Lu, Z.; Levin, L.; Ter Schure, A.F.H.; Sunderland, E.M. Total mercury released to the environment by human activities. Environ. Sci. Technol. 2017, 51, 5969–5977. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, Y.; Wang, F.; Luo, Z.; Guo, S.; Strähle, U. Toxicity of mercury: Molecular evidence. Chemosphere 2020, 245, 125586. [Google Scholar] [CrossRef] [PubMed]
- Tagliafierro, L.; Officioso, A.; Sorbo, S.; Basile, A.; Manna, C. The protective role of olive oil hydroxytyrosol against oxidative alterations induced by mercury in human erythrocytes. Food Chem. Toxicol. 2015, 82, 59–63. [Google Scholar] [CrossRef]
- Spiller, H.A. Rethinking mercury: The role of selenium in the pathophysiology of mercury toxicity. Clin. Toxicol. 2017, 56, 313–326. [Google Scholar] [CrossRef]
- Rojas, L.A.; Yañez, C.; González, M.; Lobos, S.; Smalla, K.; Seeger, M. Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE 2011, 6, e17555. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Sagasti, M.; Marino, D. PGPRs and nitrogen-fixing legumes: A perfect team for efficient Cd phytoremediation? Front. Plant Sci. 2015, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ojuederie, O.; Babalola, O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, G.; Vega-Celedón, P.; Gentina, J.C.; Seeger, M. Effects of mercury II on Cupriavidus metallidurans strain MSR33 during mercury bioremediation under aerobic and anaerobic conditions. Processes 2020, 8, 893. [Google Scholar] [CrossRef]
- Ranjard, L.; Lignier, L.; Chaussod, R. Cumulative effects of short-term polymetal contamination on soil bacterial community structure. Appl. Environ. Microbiol. 2006, 72, 1684–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Li, G.; Bishopp, A.; Heenatigala, P.P.M.; Hu, S.; Chen, Y.; Wu, Z.; Kumar, S.; Duan, P.; Yao, L.; et al. A comparison of growth on mercuric chloride for three Lemnaceae species reveals differences in growth dynamics that effect their suitability for use in either monitoring or remediating ecosystems contaminated with mercury. Front. Chem. 2018, 6, 212. [Google Scholar] [CrossRef]
- Hernández, M.; Morgante, V.; Ávila, M.; Villalobos, P.; Millares, P.; González, M.; Seeger, M. Novel s-triazine-degrading bacteria isolated from agricultural soils of Central Chile for herbicide bioremediation. Electron. J. Biotechnol. 2008, 11, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Seeger, M.; Hernández, M.; Mendez, V.; Ponce, B.; Córdova, M.; González, M. Bacterial degradation and bioremediation of chlorinated herbicides and biphenyls. J. Soil Sci. Plant Nutr. 2010, 10, 320–332. [Google Scholar] [CrossRef]
- Fuentes, S.; Barra, B.; Caporaso, J.G.; Seeger, M. From rare to dominant: A fine-tuned soil bacterial bloom during petroleum hydrocarbon bioremediation. Appl. Environ. Microbiol. 2015, 82, 888–896. [Google Scholar] [CrossRef] [Green Version]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the frontiers of life: Extremophiles in Chile and their potential for bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Deckwer, W.-D.; Becker, F.U.; Ledakowicz, S.; Wagner-Döbler, I. Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ. Sci. Technol. 2004, 38, 1858–1865. [Google Scholar] [CrossRef]
- Mahbub, K.; Krishnan, K.; Megharaj, M.; Naidu, R. Bioremediation potential of a highly mercury resistant bacterial strain Sphingobium SA2 isolated from contaminated soil. Chemosphere 2016, 144, 330–337. [Google Scholar] [CrossRef]
- McCarthy, D.; Edwards, G.C.; Gustin, M.S.; Care, A.; Miller, M.B.; Sunna, A. An innovative approach to bioremediation of mercury contaminated soils from industrial mining operations. Chemosphere 2017, 184, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Z.; Luo, H.; Zhang, M.; Zhang, D.; Pan, X.; Gadd, G.M. Multiple-pathway remediation of mercury contamination by a versatile selenite-reducing bacterium. Sci. Total Environ. 2018, 615, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Dash, H.R.; Das, S. Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegrad. 2012, 75, 207–213. [Google Scholar] [CrossRef]
- Millacura, F.A.; Janssen, P.J.; Monsieurs, P.; Janssen, A.; Provoost, A.; Van Houdt, R.; Rojas, L.A. Unintentional genomic changes endow Cupriavidus metallidurans with an augmented heavy-metal resistance. Genes 2018, 9, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero-Silva, F.; Durán, N.; Seeger, M. Synthesis of extracellular gold nanoparticles using Cupriavidus metallidurans CH34 cells. IET Nanobiotechnol. 2018, 12, 40–46. [Google Scholar] [CrossRef]
- Espinoza-Tofalos, A.; Daghio, M.; González, M.; Papacchini, M.; Franzetti, A.; Seeger, M. Toluene degradation by Cupriavidus metallidurans CH34 in nitrate-reducing conditions and in bioelectrochemical systems. FEMS Microbiol. Lett. 2018, 365, fny119. [Google Scholar] [CrossRef]
- Alviz-Gazitua, P.; Fuentes-Alburquenque, S.; Rojas, L.A.; Turner, R.; Guiliani, N.; Seeger, M. The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 2019, 10, 1499. [Google Scholar] [CrossRef]
- Seeger, M.; Rojas, L.; González, M.; Yáñez, C. Recombinant Bacterium Capable of Removing Mercury (Ii) Species, Cadmium (Ii) And Copper (Ii) In Presence of Other Heavy Metals from Polluted Sites, Product for The Bioremediation, Process of Obtaining the Product and Method of Bioremediation. U.S. Patent 8,846,376B2, 30 September 2014. [Google Scholar]
- Okino, S.; Iwasaki, K.; Yagi, O.; Tanaka, H. Development of a biological mercury removal-recovery system. Biotechnol. Lett. 2000, 22, 783–788. [Google Scholar] [CrossRef]
- Głuszcz, P.; Petera, J.; Ledakowicz, S. Mathematical modeling of the integrated process of mercury bioremediation in the industrial bioreactor. Bioproc. Biosyst. Eng. 2010, 34, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Duan, Y.; Dong, J.; Shen, S.; Si, G.; He, F.; Yang, Q.; Chen, J. Bioremediation of Hg-contaminated soil by combining a novel Hg-volatilizing Lecythophora sp. fungus, DC-F1, with biochar: Performance and the response of soil fungal community. Sci. Total Environ. 2019, 671, 676–684. [Google Scholar] [CrossRef]
- Voijant, B.T.; Rozaimah, S.S.A.; Basri, H.; Idris, M.; Anuar, N.; Mukhlisin, M. A review on heavy metals (As, Pb, and Hg) uptake by plants through phytoremediation. Int. J. Chem. Eng. 2011, 2011, 1–31. [Google Scholar] [CrossRef]
- Zgorelec, Z.; Bilandzija, N.; Knez, K.; Galic, M.; Zuz, S. Cadmium and mercury phytostabilization from soil using Miscanthus × giganteus. Sci. Rep. 2020, 10, 6685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, M.R.; Banerjee, D.K.; Fedorak, P.M.; Hashimoto, A.; Masliyah, J.H.; Pickard, M.A. Biological remediation of anthracene-contaminated soil in rotating bioreactors. Appl. Microbiol. Biot. 1994, 40, 933–940. [Google Scholar] [CrossRef]
- Banerjee, D.K.; Fedorak, P.M.; Hashimoto, A.; Masliyah, J.H.; Pickard, M.A.; Gray, M.R. Monitoring the biological treatment of anthracene-contaminated soil in a rotating-drum bioreactor. Appl. Microbiol. Biotechnol. 1995, 43, 521–528. [Google Scholar] [CrossRef]
- Pinelli, D.; Fava, F.; Nocentini, M.; Pasquali, G. Bioremediation of a polycyclic aromatic hydrocarbon-contaminated soil by using different aerobic batch bioreactor systems. Soil Sediment. Contam. 1997, 6, 243–256. [Google Scholar] [CrossRef]
- Woo, S.H.; Park, J.M. Evaluation of drum bioreactor performance used for decontamination of soil polluted with polycyclic aromatic hydrocarbons. J. Chem. Technol. Biotechnol. 1999, 74, 937–944. [Google Scholar] [CrossRef]
- Hardin, M.T.; Howes, T.; Mitchell, D.A. Mass transfer correlations for rotating drum bioreactors. J. Biotechnol. 2002, 97, 89–101. [Google Scholar] [CrossRef]
- Barrera-Cortés, J.; Manilla-Pérez, E.; Poggi-Varaldo, H.M. Oxygen transfer to slurries treated in a rotating drum operated at atmospheric pressure. Bioproc. Biosyst. Eng. 2006, 29, 391–398. [Google Scholar] [CrossRef]
- Rodríguez-Meza, M.A.; Chávez-Gómez, B.; Poggi-Varaldo, H.M.; Ríos-Leal, E.; Barrera-Cortés, J. Design of a new rotating drum bioreactor operated at atmospheric pressure on the bioremediation of a polluted soil. Bioproc. Biosyst. Eng. 2009, 33, 573–582. [Google Scholar] [CrossRef]
- Jaramillo, A.C.; Cobas, M.; Hormaza, A.; Sanromá, M.A. Degradation of adsorbed azo dye by solid-state fermentation: Improvement of culture conditions, a kinetic study, and rotating drum bioreactor performance. Water Air Soil Pollut. 2017, 228, 205. [Google Scholar] [CrossRef]
- Jin, J.; Liu, G.; Shi, S.; Cong, W. Studies on the performance of a rotating drum bioreactor for bioleaching processes—Oxygen transfer, solids distribution and power consumption. Hydrometallurgy 2010, 103, 30–34. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rösch, C.; Mergel, A.; Bothe, H. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 2002, 68, 3818–3829. [Google Scholar] [CrossRef] [Green Version]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AOAC Official Method 977.15 Mercury in Fish Alternative Flameless Atomic Absorption Spectrophotometric Method. First Action 1977 Final Action 1978. Available online: http://www.aoacofficialmethod.org/index.php?main_page=product_info&cPath=1&products_id=2383 (accessed on 14 November 2020).
- Morgante, V.; López-López, A.; Flores, C.; González, M.; González, B.; Vásquez, V.; Rossello-Mora, R.; Seeger, M. Bioaugmentation with Pseudomonas sp. strain MHP41 promotes simazine attenuation and bacterial community changes in agricultural soils. FEMS Microbiol. Ecol. 2010, 71, 114–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Brankatschk, R.; Bodenhausen, N.; Zeyer, J.; Bürgmann, H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl. Environ. Microbiol. 2012, 78, 4481–4489. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4789. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Rineau, F.; Weyens, N.; Vangronsveld, J. Towards an enhanced understanding of plant-microbiome interactions to improve phytoremediation: Engineering the metaorganism. Front. Microbiol. 2016, 16, 341. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Weyens, N.; Vangronsveld, J. Phytoremediation: State-of-the-art and a key role for the plant microbiome in future trends and research prospects. Int. J. Phytoremediat. 2017, 19, 23–38. [Google Scholar] [CrossRef]
- Mariano, C.; Mello, I.S.; Barros, B.M.; da Silva, G.F.; Terezo, A.J.; Soares, M.A. Mercury alters the rhizobacterial community in Brazilian wetlands and it can be bioremediated by the plant-bacteria association. Environ. Sci. Pollut. Res. Int. 2020, 27, 13550–13564. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, D.; Cao, Y.; Sik Ok, Y.; Tack, F.M.; Rinklebe, J.; O’Connor, D. Remediation of mercury contaminated soil, water, and air: A review of emerging materials and innovative technologies. Environ. Int. 2020, 134, 105281. [Google Scholar] [CrossRef] [PubMed]
- Asztalos, E.; Sipka, G.; Kis, M.; Trotta, M.; Maróti, P. The reaction center is the sensitive target of the mercury (II) ion in intact cells of photosynthetic bacteria. Photosynth. Res. 2012, 112, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Branco, V.; Ramos, P.; Canario, J.; Lu, J.; Holmgren, A.; Carvalho, C. Biomarkers of adverse response to mercury: Histopathology versus thioredoxin reductase activity. J. Biomed. Biotechnol. 2012, 2012, 1–9. [Google Scholar] [CrossRef]
- Wang, Y.; Robison, T.; Wiatrowski, H. The impact of ionic mercury on antioxidant defenses in two mercury-sensitive anaerobic bacteria. BioMetals 2013, 26, 1023–1031. [Google Scholar] [CrossRef]
- Pepi, M.; Lobianco, A.; Renzi, M.; Perra, G.; Bernardini, E.; Marvasi, M.; Gasperini, S.; Volterrani, M.; Franchi, E.; Heipieper, H.J.; et al. Two naphthalene degrading bacteria belonging to the genera Paenibacillus and Pseudomonas isolated from a highly polluted lagoon perform different sensitivities to the organic and heavy metal contaminants. Extremophiles 2009, 13, 839–848. [Google Scholar] [CrossRef]
- Rasmussen, L.D.; Zawadsky, C.; Binnerup, S.J.; Øregaard, G.; Sørensen, S.J.; Kroer, N. Cultivation of hard-to-culture subsurface mercury-resistant bacteria and discovery of new merA gene sequences. Appl. Environ. Microbiol. 2008, 74, 3795–3803. [Google Scholar] [CrossRef] [Green Version]
- Lapanje, A.; Zrimec, A.; Drobne, D.; Rupnik, M. Long-term Hg pollution-induced structural shifts of bacterial community in the terrestrial isopod (Porcellio scaber) gut. Environ. Pollut. 2010, 158, 3186–3193. [Google Scholar] [CrossRef]
- François, F.; Lombard, C.; Guigner, J.M.; Soreau, P.; Brian-Jaisson, F.; Martino, G.; Vandervennet, M.; Garcia, D.; Molinier, A.L.; Pignol, D.; et al. Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl. Environ. Microbiol. 2012, 78, 1097–1106. [Google Scholar] [CrossRef] [Green Version]
- Chasanah, U.; Nuraini, Y.; Handayanto, E. The potential of mercury-resistant bacteria isolated from small-scale gold mine tailings for accumulation of mercury. Ecol. Eng. 2018, 19, 236–245. [Google Scholar] [CrossRef]
- Naguib, M.M.; Khairalla, A.S.; El-Gendy, A.O.; Elkhatib, W.F. Isolation and characterization of mercury-resistant bacteria from wastewater sources in Egypt. Can. J. Microbiol. 2019, 65, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Tiwary, B.N. Isolation of a novel strain of Planomicrobium chinense from diesel contaminated soil of tropical environment. J. Basic Microbiol. 2013, 53, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Duraisamy, P.; Sekar, J.; Arunkumar, A.D.; Ramalingam, P.V. Kinetics of phenol biodegradation by heavy metal tolerant rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from distillery effluent contaminated soils. Front. Microbiol. 2020, 11, 1573. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.G.; Hurtado, R.; Gomes, L.G.R.; Profeta, R.; Rifici, C.; Attili, A.R.; Spier, S.J.; Giuseppe, M.; Morais-Rodrigues, F.; Gomide, A.C.P.; et al. Complete genome analysis of Glutamicibacter creatinolyticus from mare abscess and comparative genomics provide insight of diversity and adaptation for Glutamicibacter. Gene 2020, 741, 144566. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.K.; Barkay, T.; Al-Soud, W.A.; Sørensen, S.J.; Skov, H.; Kroer, N. Diversity and characterization of mercury-resistant bacteria in snow, freshwater and sea-ice brine from the High Arctic. FEMS Microbiol. Ecol. 2011, 75, 390–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Medina, J.A.; Enriquez, J.; Cruz, A.; Garcia, R.; Solis, S.; Hernandez, G.; Jones, G.H.; Campos-Guillen, J. Isolation and characterization of mercury resistant Bacillus sp. from soils with an extensive history as substrates for mercury extraction in Mexico. Geomicrobiol. J. 2013, 30, 454–461. [Google Scholar] [CrossRef]
- Figueiredo, N.L.; Areias, A.; Mendes, R.; Canário, J.; Duarte, A.; Carvalho, C. Mercury-resistant bacteria from salt marsh of Tagus estuary: The influence of plants presence and mercury contamination levels. J. Toxicol. Environ. 2014, 77, 959–971. [Google Scholar] [CrossRef]
- Boyd, E.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef] [Green Version]
- Smalla, K.; Jechalke, S.; Top, E.M. Plasmid detection, characterization and ecology. Microbiol. Spectr. 2015, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Smalla, K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012, 6, 1083–1104. [Google Scholar] [CrossRef] [Green Version]
- Smalla, K.; Haines, A.S.; Jones, K.; Krogerrecklenfort, E.; Heuer, H.; Schloter, M.; Thomas, C.M. Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury polluted river sediments-first discovery of IncP-1β plasmids with a complex mer transposon as sole accessory element. Appl. Environ. Microbiol. 2006, 72, 7253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musovic, S.; Oregaard, G.; Kroer, N.; Sørensen, S.J. Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among gram-positive and gram-negative bacteria indigenous to the barley rhizosphere. Appl. Environ. Microbiol. 2006, 80, 6687–6692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shintani, M.; Matsui, K.; Inoue, J.; Hosoyama, A.; Ohji, S.; Yamazoe, A.; Nojiri, H.; Kimbara, K.; Ohkuma, M. Single-cell analyses revealed transfer ranges of IncP-1, IncP-7, and IncP-9 plasmids in a soil bacterial community. Appl. Environ. Microbiol. 2013, 80, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.; Sørensen, S.; Smets, B. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015, 9, 934–945. [Google Scholar] [CrossRef] [Green Version]
- Smets, B.F.; Morrow, J.B.; Arango Pinedo, C. Plasmid introduction in metal-stressed, subsurface-derived microcosms: Plasmid fate and community response. Appl. Environ. Microbiol. 2003, 69, 4087–4097. [Google Scholar] [CrossRef] [Green Version]
- Garbisu, C.; Garaiyurrebaso, O.; Epelde, L.; Grohmann, E.; Alkorta, I. Plasmid-mediated bioaugmentation for the bioremediation of contaminated soils. Front. Microbiol. 2017, 8, 1966. [Google Scholar] [CrossRef] [Green Version]
- Matsui, K.; Endo, G. Mercury bioremediation by mercury resistance transposon-mediated in situ molecular breeding. Appl. Microbiol. Biotechnol. 2018, 102, 3037–3048. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Y.; Shen, J.; Zhang, L.; He, J. Effects of mercury on the activity and community composition of soil ammonia oxidizers. Environ. Sci. Pollut. R. 2010, 17, 1237–1244. [Google Scholar] [CrossRef] [Green Version]
- van Dorst, J.; Siciliano, S.D.; Winsley, T.; Snape, I.; Ferrari, B.C. Bacterial targets as potential indicators of diesel fuel toxicity in subantarctic soils. Appl. Environ. Microbiol. 2014, 80, 4021–4033. [Google Scholar] [CrossRef] [Green Version]
- Levy-Booth, D.J.; Prescott, C.E.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Ke, X.; Lu, W.; Conrad, R. High oxygen concentration increases the abundance and activity of bacterial rather than archaeal nitrifiers in rice field soil. Microb. Ecol. 2015, 70, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Yeom, J.; Kim, J.; Han, J.; Soo, H.L.; Park, H.; Hyun, S.; Park, W. Change in gene abundance in the nitrogen biogeochemical cycle with temperature and nitrogen addition in Antarctic soils. Res. Microbiol. 2011, 162, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Franche, C.; Lindström, K.; Elmerich, C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant. Soil 2008, 321, 35–59. [Google Scholar] [CrossRef]
- Lladó, S.; Grácia, E.; Solanas, A.M.; Viñas, M. Fungal and bacterial microbial community assessment during bioremediation assays in an aged creosote-polluted soil. Soil Biol. Biochem. 2013, 67, 114–123. [Google Scholar] [CrossRef] [Green Version]








| Primers | Sequence (5′-3′) | Gene Target | Size (pb) | Reference |
|---|---|---|---|---|
| 27F | AGAGTTTGATCMTGGCTCAG | 16S rRNA | 1465 | [53] |
| 1492R | TACGGYTACCTTGTTACGACTT | |||
| nifH-F-Rösch | AAAGGYGGWATCGGYAARTCCACCA | nifH | 458 | [54] |
| nifH-R-Rösch | TTGTTSGCSGCRTACATSGCCATCAT | |||
| amoA-1F | GGGGTTTCTACTGGTGGT | AOB amoA | 491 | [55] |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | |||
| zniA_F | GGAAAGGCCTTCCTGGACAT | zniA | 167 | L. Rojas, personal communication |
| zniA_R | TCAACGCGGAGTTCTTCGTA | |||
| merG_F | AGTACCGCAACGTTAGGCAT | merG | 171 | L. Rojas, personal communication |
| merG_R | ACCGCATTTGTACGCAAGAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo, G.; Vega-Celedón, P.; Gentina, J.C.; Seeger, M. Bioremediation by Cupriavidus metallidurans Strain MSR33 of Mercury-Polluted Agricultural Soil in a Rotary Drum Bioreactor and Its Effects on Nitrogen Cycle Microorganisms. Microorganisms 2020, 8, 1952. https://doi.org/10.3390/microorganisms8121952
Bravo G, Vega-Celedón P, Gentina JC, Seeger M. Bioremediation by Cupriavidus metallidurans Strain MSR33 of Mercury-Polluted Agricultural Soil in a Rotary Drum Bioreactor and Its Effects on Nitrogen Cycle Microorganisms. Microorganisms. 2020; 8(12):1952. https://doi.org/10.3390/microorganisms8121952
Chicago/Turabian StyleBravo, Guillermo, Paulina Vega-Celedón, Juan Carlos Gentina, and Michael Seeger. 2020. "Bioremediation by Cupriavidus metallidurans Strain MSR33 of Mercury-Polluted Agricultural Soil in a Rotary Drum Bioreactor and Its Effects on Nitrogen Cycle Microorganisms" Microorganisms 8, no. 12: 1952. https://doi.org/10.3390/microorganisms8121952
APA StyleBravo, G., Vega-Celedón, P., Gentina, J. C., & Seeger, M. (2020). Bioremediation by Cupriavidus metallidurans Strain MSR33 of Mercury-Polluted Agricultural Soil in a Rotary Drum Bioreactor and Its Effects on Nitrogen Cycle Microorganisms. Microorganisms, 8(12), 1952. https://doi.org/10.3390/microorganisms8121952