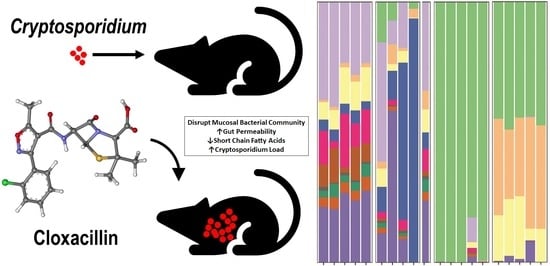
Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oocyst Preparation
2.2. Cryptosporidium Parvum Mouse Infection and Antibiotic Treatment
2.3. DNA Extraction, Polymerase Chain Reaction (PCR), Sequencing, and Sequence Analysis
2.4. Bioinformatics and Statistical Analysis
2.5. Short-Chain Fatty Acids Determination
2.6. Gut Permeability Assay
2.7. Statistical Analysis for Parasite Load and Fluorescein Isothiocyanate (FITC)-Dextran Permeability Assay
3. Results
3.1. Microbiota and Infection Levels Are Altered in Response to Antibiotic Treatment
3.2. Gut Permeability Is Increased in Infected Mice
3.3. Short-Chain Fatty Acids Are Decreased in Antibiotic Treated Mice
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Shirley, D.A.; Moonah, S.N.; Kotloff, K.L. Burden of disease from cryptosporidiosis. Curr. Opin. Infect. Dis. 2012, 25, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Checkley, W.; White, A.C., Jr.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Painter, J.E.; Hlavsa, M.C.; Collier, S.A.; Xiao, L.; Yoder, J.S.; Centers for Disease Control and Prevention. Cryptosporidiosis surveillance -- United States, 2011–2012. MMWR Suppl. 2015, 64, 1–14. [Google Scholar] [PubMed]
- Gilchrist, C.A.; Petri, S.E.; Schneider, B.N.; Reichman, D.J.; Jiang, N.; Begum, S.; Watanabe, K.; Jansen, C.S.; Elliott, K.P.; Burgess, S.L.; et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J. Infect. Dis. 2016, 213, 1579–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barash, N.R.; Maloney, J.G.; Singer, S.M.; Dawson, S.C. Giardia alters commensal microbial diversity throughout the murine gut. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarino, N.F.; LeCleir, G.R.; Denny, J.E.; Dearth, S.P.; Harding, C.L.; Sloan, S.S.; Gribble, J.L.; Campagna, S.R.; Wilhelm, S.W.; Schmidt, N.W. Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. USA 2016, 113, 2235–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partida-Rodriguez, O.; Serrano-Vazquez, A.; Nieves-Ramirez, M.E.; Moran, P.; Rojas, L.; Portillo, T.; Gonzalez, E.; Hernandez, E.; Finlay, B.B.; Ximenez, C. Human intestinal microbiota: Interaction between parasites and the host immune response. Arch. Med. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chappell, C.L.; Darkoh, C.; Shimmin, L.; Farhana, N.; Kim, D.K.; Okhuysen, P.C.; Hixson, J. Fecal indole as a biomarker of susceptibility to Cryptosporidium infection. Infect. Immun. 2016, 84, 2299–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harp, J.A.; Wannemuehler, M.W.; Woodmansee, D.B.; Moon, H.W. Susceptibility of germfree of antibiotic-treated adult mice to Cryptosporidium parvum. Infect. Immun. 1988, 56, 2006–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, R.E.; Haynes, J.S.; Harp, J.A.; Waters, W.R.; Wannemuehler, M.J. Cryptosporidium parvum initiates inflammatory bowel disease in germfree T cell receptor-alpha-deficient mice. Am. J. Pathol. 1998, 153, 1717–1722. [Google Scholar] [CrossRef]
- Lantier, L.; Drouet, F.; Guesdon, W.; Mancassola, R.; Metton, C.; Lo-Man, R.; Werts, C.; Laurent, F.; Lacroix-Lamande, S. Poly(I:C)-induced protection of neonatal mice against intestinal Cryptosporidium parvum infection requires an additional TLR5 signal provided by the gut flora. J. Infect. Dis. 2014, 209, 457–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorla, S.K.; McNair, N.N.; Yang, G.; Gao, S.; Hu, M.; Jala, V.R.; Haribabu, B.; Striepen, B.; Cuny, G.D.; Mead, J.R.; et al. Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob. Agents Chemother. 2014, 58, 1603–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrowood, M.J.; Donaldson, K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 1996, 43, S89. [Google Scholar] [CrossRef] [PubMed]
- Arrowood, M.J.; Sterling, C.R. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J. Parasitol. 1987, 73, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Arrowood, M.J.; Hurd, M.R.; Mead, J.R. A new method for evaluating experimental cryptosporidial parasite loads using immunofluorescent flow cytometry. J. Parasitol. 1995, 81, 404–409. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, G.; Nyman, M.; Jonsson, J.A. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed. Chromatogr. 2006, 20, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Kolachala, V.; Dalmasso, G.; Nguyen, H.; Laroui, H.; Sitaraman, S.V.; Merlin, D. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS ONE 2009, 4, e6073. [Google Scholar] [CrossRef] [PubMed]
- Leys, C.; Ley, C.; Klein, O.; Bernard, P.; Licata, L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 2013, 49, 764–766. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.J.; Flanigan, T.P. Paromomycin inhibits Cryptosporidium infection of a human enterocyte cell line. J. Infect. Dis. 1992, 165, 772–774. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhang, H.; Fritzler, J.M.; Rider, S.D., Jr.; Xiang, L.; McNair, N.N.; Mead, J.R.; Zhu, G. Amelioration of Cryptosporidium parvum infection in vitro and in vivo by targeting parasite fatty acyl-coenzyme A synthetases. J. Infect. Dis. 2014, 209, 1279–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Ras, R.; Huynh, K.; Desoky, E.; Badawy, A.; Widmer, G. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int. J. Parasitol. 2015, 45, 567–573. [Google Scholar] [CrossRef] [PubMed]
- McKenney, E.A.; Greene, L.K.; Drea, C.M.; Yoder, A.D. Down for the count: Cryptosporidium infection depletes the gut microbiome in Coquerel’s sifakas. Microb. Ecol. Health Dis. 2017, 28, 1335165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abera, B.; Hailu, T.; Beza, L.; Mulu, W.; Yizengaw, E.; Kibret, M. Aetiology of acute diarrhoea and antimicrobial usage among children aged under five years at health centres in Bahir Dar, Ethiopia. Trop. Dr. 2020, 0049475520912558. [Google Scholar] [CrossRef] [PubMed]
- Fritzler, J.M.; Zhu, G. Novel anti-Cryptosporidium activity of known drugs identified by high-throughput screening against parasite fatty acyl-CoA binding protein (ACBP). J. Antimicrob. Chemother. 2012, 67, 609–617. [Google Scholar] [CrossRef]
- Craven, M.; Egan, C.E.; Dowd, S.E.; McDonough, S.P.; Dogan, B.; Denkers, E.Y.; Bowman, D.; Scherl, E.J.; Simpson, K.W. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS ONE 2012, 7, e41594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choo, J.M.; Kanno, T.; Zain, N.M.; Leong, L.E.; Abell, G.C.; Keeble, J.E.; Bruce, K.D.; Mason, A.J.; Rogers, G.B. Divergent Relationships between Fecal Microbiota and Metabolome following Distinct Antibiotic-Induced Disruptions. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, B.C.M.; Widmer, G. Probiotic Product Enhances Susceptibility of Mice to Cryptosporidiosis. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. BioMed Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, V.; Korbel, D.S.; Barakat, F.M.; Choudhry, N.; Petry, F. Innate immune responses against Cryptosporidium parvum infection. Parasite Immunol. 2013, 35, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, K.; Chen, H.; Su, Y.; Zhu, W. Caecal infusion of the short-chain fatty acid propionate affects the microbiota and expression of inflammatory cytokines in the colon in a fistula pig model. Microb. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, O.P.; Ranganna, K.; Yatsu, F.M. Butyrate, an HDAC inhibitor, stimulates interplay between different posttranslational modifications of histone H3 and differently alters G1-specific cell cycle proteins in vascular smooth muscle cells. Biomed. Pharmacother. 2010, 64, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Bedi, B.; McNair, N.N.; Forster, I.; Mead, J.R. Il-18 cytokine levels modulate innate immune responses and cryptosporidiosis in mice. J. Eukaryot. Microbiol. 2015, 62, 44–50. [Google Scholar] [CrossRef] [PubMed]
- McDonald, V.; Pollok, R.C.; Dhaliwal, W.; Naik, S.; Farthing, M.J.; Bajaj-Elliott, M. A potential role for interleukin-18 in inhibition of the development of Cryptosporidium parvum. Clin. Exp. Immunol. 2006, 145, 555–562. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, J.K.; Moore, R.; Dooley, S.; Keusch, G.T.; Tzipori, S. Cryptosporidium parvum infection of Caco-2 cell monolayers induces an apical monolayer defect, selectively increases transmonolayer permeability, and causes epithelial cell death. Infect. Immun. 1994, 62, 4506–4514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Chatterjee, I.; Anbazhagan, A.N.; Jayawardena, D.; Priyamvada, S.; Alrefai, W.A.; Sun, J.; Borthakur, A.; Dudeja, P.K. Cryptosporidium parvum disrupts intestinal epithelial barrier function via altering expression of key tight junction and adherens junction proteins. Cell. Microbiol. 2018, 20, e12830. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef] [PubMed] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charania, R.; Wade, B.E.; McNair, N.N.; Mead, J.R. Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels. Microorganisms 2020, 8, 879. https://doi.org/10.3390/microorganisms8060879
Charania R, Wade BE, McNair NN, Mead JR. Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels. Microorganisms. 2020; 8(6):879. https://doi.org/10.3390/microorganisms8060879
Chicago/Turabian StyleCharania, Raheela, Brandy E. Wade, Nina N. McNair, and Jan R. Mead. 2020. "Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels" Microorganisms 8, no. 6: 879. https://doi.org/10.3390/microorganisms8060879
APA StyleCharania, R., Wade, B. E., McNair, N. N., & Mead, J. R. (2020). Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels. Microorganisms, 8(6), 879. https://doi.org/10.3390/microorganisms8060879