Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture
Abstract
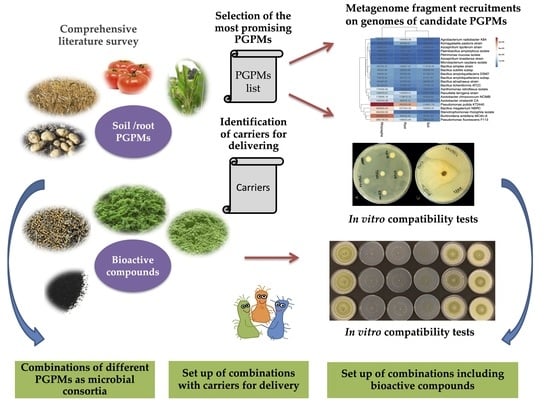
:1. Introduction
2. Materials and Methods
2.1. Literature Survey: Search Strategy and Data Collection
2.2. Metagenome Fragment Recruitments on Genomes of Candidate PGPM
| Algorithm 1: Filter Recruited Reads that Are Located at Highly Covered Regions |
| Input: recruited reads R Output: qualified reads Q
|
2.3. Microbe-Microbe In Vitro Compatibility Test
2.4. Effects of Bioactive Compounds on Microbial Growth In Vitro
3. Results and Discussion
3.1. Identification of the Most Promising Beneficial Microorganisms and Carriers
3.2. Metagenome Fragment Recruitments on Genomes of Candidate PGP Soil Microorganisms Represented in Databases
3.3. Evaluation of In Vitro Co-Culture Compatibility of Selected Microbial Strains
3.4. Design of Microbial Consortia Inoculants
3.5. Prebiotic Effect of the Bioactive Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil Beneficial Bacteria and Their Role in Plant Growth Promotion: A Review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant Growth-Promoting Bacteria in the Rhizo- and Endosphere of Plants: Their Role, Colonization, Mechanisms Involved and Prospects for Utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.L.; Pepe, O. Microbial Consortia: Promising Probiotics as Plant Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1801. [Google Scholar] [CrossRef]
- Kim, Y.C.; Anderson, A.J. Rhizosphere Pseudomonads as Probiotics Improving Plant Health. Mol. Plant Pathol. 2018, 19, 2349–2359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maheshwari, D.K. Bacteria in Agrobiology: Plant Probiotics; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability-A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- Berg, G. Plant-Microbe Interactions Promoting Plant Growth and Health: Perspectives for Controlled Use of Microorganisms in Agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Enebe, M.C.; Babalola, O. The Impact of Microbes in the Orchestration of Plants’ Resistance to Biotic Stress: A Disease Management Approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural Uses of Plant Biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef] [Green Version]
- Walker, R.; Otto-Pille, C.; Gupta, S.; Schillaci, M.; Roessner, U. Current Perspectives and Applications in Plant Probiotics. Microbiol. Aust. 2020, 41, 95–99. [Google Scholar] [CrossRef]
- Bashan, Y. Inoculants of Plant Growth-Promoting Bacteria for Use in Agriculture. Biotechnol. Adv. 1998, 16, 729–770. [Google Scholar] [CrossRef]
- Bevivino, A. Field Microbial Application to Foster Food Quality and Safety. SIMBA Project. 2020. Available online: http://simbaproject.eu/field-microbial-application-to-foster-food-quality-and-safety/ (accessed on 5 June 2020).
- Ambrosini, A.; de Souza, R.; Passaglia, L.M.P. Ecological Role of Bacterial Inoculants and Their Potential Impact on Soil Microbial Diversity. Plant Soil 2016, 400, 193–207. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and Challenges of Microbial Application for Crop Improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.P. Advances in Plant Growth-Promoting Bacterial Inoculant Technology: Formulations and Practical Perspectives (1998-2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Bradáčová, K.; Florea, A.S.; Bar-Tal, A.; Minz, D.; Yermiyahu, U.; Shawahna, R.; Kraut-Cohen, J.; Zolti, A.; Erel, R.; Dietel, K.; et al. Microbial Consortia versus Single-Strain Inoculants: An Advantage in PGPM-Assisted Tomato Production? Agronomy 2019, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Kumar, N.; Shandilya, C.; Mohapatra, S.; Bhayana, S.; Varma, A. Revisiting Plant–Microbe Interactions and Microbial Consortia Application for Enhancing Sustainable Agriculture: A Review. Front. Microbiol. 2020, 11, 3195. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A Review on the Plant Microbiome: Ecology, Functions, and Emerging Trends in Microbial Application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Khare, E.; Arora, N.K. Effects of Soil Environment on Field Efficacy of Microbial Inoculants. In Plant Microbes Symbiosis: Applied Facets; Springer: New Delhi, India, 2015. [Google Scholar]
- Raeid, M.M.; Al-Kharusi, S.; Al-Hinai, M. Effect of Biostimulation, Temperature and Salinity on Respiration Activities and Bacterial Community Composition in an Oil Polluted Desert Soil. Int. Biodeterior. Biodegrad. 2015, 98, 42–52. [Google Scholar]
- Bargaz, A.; Lyamlouli, K.; Chtouki, M.; Zeroual, Y.; Dhiba, D. Soil Microbial Resources for Improving Fertilizers Efficiency in an Integrated Plant Nutrient Management System. Front. Microbiol. 2018, 9, 1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelkner, J.; Henke, C.; Lin, T.W.; Pätzold, W.; Hassa, J.; Jaenicke, S.; Grosch, R.; Pühler, A.; Sczyrba, A.; Schlüter, A. Effect of Long-Term Farming Practices on Agricultural Soil Microbiome Members Represented by Metagenomically Assembled Genomes (MAGs) and Their Predicted Plant-Beneficial Genes. Genes 2019, 10, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kröber, M.; Wibberg, D.; Grosch, R.; Eikmeyer, F.; Verwaaijen, B.; Chowdhury, S.P.; Hartmann, A.; Pühler, A.; Schlüter, A. Effect of the Strain Bacillus Amyloliquefaciens FZB42 on the Microbial Community in the Rhizosphere of Lettuce under Field Conditions Analyzed by Whole Metagenome Sequencing. Front. Microbiol. 2014, 5, 252. [Google Scholar]
- Rouphael, Y.; Colla, G. Editorial: Biostimulants in Agriculture. Front. Plant Sci. 2020, 11, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouphael, Y.; Colla, G. Synergistic Biostimulatory Action: Designing the next Generation of Plant Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Communication from the Commission of the European Parliament, the Council, the European economic and social committee and the committee of the regions. A Farm to Fork Strategy: For a Fair, Healthy and Environmental-Friendly Food System. 2020. Brussels COM(2020) 381 Final. Available online: https://ec.europa.eu/info/sites/info/files/communication-annex-farm-fork-green-deal_en.pdf (accessed on 4 October 2020).
- Colla, G.; Rouphael, Y.; Canaguier, R.; Svecova, E.; Cardarelli, M. Biostimulant Action of a Plant-Derived Protein Hydrolysate Produced through Enzymatic Hydrolysis. Front. Plant Sci. 2014, 5, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Švecová, E.B.; Ruzzi, M. Foliar Application of Vegetal-Derived Bioactive Compounds Stimulates the Growth of Beneficial Bacteria and Enhances Microbiome Biodiversity in Lettuce. Front. Plant Sci. 2019, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Francesca, S.; Arena, C.; Hay Mele, B.; Schettini, C.; Ambrosino, P.; Barone, A.; Rigano, M.M. The Use of a Plant-Based Biostimulant Improves Plant Performances and Fruit Quality in Tomato Plants Grown at Elevated Temperatures. Agronomy 2020, 10, 363. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Krüger, J.; Sczyrba, A. Analyzing Large Scale Genomic Data on the Cloud with Sparkhit. Bioinformatics 2018, 34, 1457–1465. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.; Zhu, Z.; Fu, L.; Wu, S.; Li, W. FR-HIT, a Very Fast Program to Recruit Metagenomic Reads to Homologous Reference Genomes. Bioinformatics 2011, 27, 1704–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irabor, A.; Mmbaga, M.T. Evaluation of Selected Bacterial Endophytes for Biocontrol Potential against Phytophthora Blight of Bell Pepper (Capsicum Annuum L.). J. Plant Pathol. Microbiol. 2017, 8, 10. [Google Scholar]
- Siddiqui, I.A.; Shaukat, S.S. Combination of Pseudomonas Aeruginosa and Pochonia Chlamydosporia for Control of Root-Infecting Fungi in Tomato. J. Phytopathol. 2003, 151, 215–222. [Google Scholar] [CrossRef]
- Masoero, G.; Peiretti, P.G.; Cugnetto, A.; Giovannetti, G. Raw PH Fall-out as a Sign of a Mycorrhizal Modifier of Sorghum Sudanensis. J. Agron. Res. 2018, 1, 1–11. [Google Scholar] [CrossRef]
- Holguin, G.; Bashan, Y. Nitrogen-Fixation by Azospirillum Brasilense CD Is Promoted When Co-Cultured with a Mangrove Rhizosphere Bacterium (Staphylococcus Sp.). Soil Biol. Biochem. 1996, 28, 1651–1660. [Google Scholar] [CrossRef]
- Rozier, C.; Hamzaoui, J.; Lemoine, D.; Czarnes, S.; Legendre, L. Field-Based Assessment of the Mechanism of Maize Yield Enhancement by Azospirillum Lipoferum CRT1. Sci. Rep. 2017, 7, 7416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viscardi, S.; Ventorino, V.; Duran, P.; Maggio, A.; De Pascale, S.; Mora, M.L.; Pepe, O. Assessment of Plant Growth Promoting Activities and Abiotic Stress Tolerance of Azotobacter Chroococcum Strains for a Potential Use in Sustainable Agriculture. J. Soil Sci. Plant Nutr. 2016, 16, 848–863. [Google Scholar] [CrossRef] [Green Version]
- Biari, A.; Gholami, A.; Rahmani, H.A. Growth Promotion and Enhanced Nutrient Uptake of Maize (Zea Mays L.) by Application of Plant Growth Promoting Rhizobacteria in Arid Region of Iran. J. Biol. Sci. 2008, 8, 1015–1020. [Google Scholar] [CrossRef] [Green Version]
- Gholami, A. Growth Response of Corn during Growing Season by PGPR. In Plant Growth Promotion by Rhizobacteria for Sustainable Agriculture; Reddy, M.S., Sayyed, R.Z., Sarma, Y.R., Reddy, K.R.K., Kloepper, J.W., Desai, S., Rao, V.K., Reddy, B.C., Podile, A.R., Eds.; Scientific Publishers: New Delhi, India, 2010. [Google Scholar]
- Ferreira, C.M.H.; López-Rayo, S.; Lucena, J.J.; Soares, E.V.; Soares, H.M.V.M. Evaluation of the Efficacy of Two New Biotechnological-Based Freeze-Dried Fertilizers for Sustainable Fe Deficiency Correction of Soybean Plants Grown in Calcareous Soils. Front. Plant Sci. 2019, 10, 1335. [Google Scholar] [CrossRef] [PubMed]
- Noar, J.D.; Bruno-Bárcena, J.M. Azotobacter Vinelandii: The Source of 100 Years of Discoveries and Many More to Come. Microbiology 2018, 164, 421–436. [Google Scholar] [CrossRef]
- Mpanga, I.K.; Dapaah, H.K.; Geistlinger, J.; Ludewig, U.; Neumann, G. Soil Type-Dependent Interactions of p-Solubilizing Microorganisms with Organic and Inorganic Fertilizers Mediate Plant Growth Promotion in Tomato. Agronomy 2018, 8, 213. [Google Scholar] [CrossRef] [Green Version]
- Moradtalab, N.; Ahmed, A.; Geistlinger, J.; Walker, F.; Höglinger, B.; Ludewig, U.; Neumann, G. Synergisms of Microbial Consortia, N Forms, and Micronutrients Alleviate Oxidative Damage and Stimulate Hormonal Cold Stress Adaptations in Maize. Front. Plant Sci. 2020, 11, 396. [Google Scholar] [CrossRef]
- Turan, M.; Gulluce, M.; von Wirén, N.; Sahin, F. Yield Promotion and Phosphorus Solubilization by Plant Growth-Promoting Rhizobacteria in Extensive Wheat Production in Turkey. J. Plant Nutr. Soil Sci. 2012, 175, 818–826. [Google Scholar] [CrossRef]
- Çakmakçi, R.; Kantar, F.; Sahin, F. Effect of N2-Fixing Bacterial Inoculations on Yield of Sugar Beet and Barley. J. Plant Nutr. Soil Sci. 2001, 164, 527–531. [Google Scholar] [CrossRef]
- Saeid, A.; Prochownik, E.; Dobrowolska-Iwanek, J. Phosphorus Solubilization by Bacillus Species. Molecules 2018, 23, 2897. [Google Scholar] [CrossRef] [Green Version]
- Ghyselinck, J.; Velivelli, S.L.S.; Heylen, K.; O’Herlihy, E.; Franco, J.; Rojas, M.; De Vos, P.; Prestwich, B.D. Bioprospecting in Potato Fields in the Central Andean Highlands: Screening of Rhizobacteria for Plant Growth-Promoting Properties. Syst. Appl. Microbiol. 2013, 36, 116–127. [Google Scholar] [CrossRef]
- Yao, A.; Bochow, H.; Karimov, S.; Boturov, U.; Sanginboy, S.; Sharipov, A. Effect of FZB 24® Bacillus Subtilis as a Biofertilizer on Cotton Yields in Field Tests. Arch. Phytopathol. Plant Prot. 2006, 39, 323–328. [Google Scholar] [CrossRef]
- Mpanga, I.K.; Nkebiwe, P.M.; Kuhlmann, M.; Cozzolino, V.; Piccolo, A.; Geistlinger, J.; Berger, N.; Ludewig, U.; Neumann, G. The Form of n Supply Determines Plant Growth Promotion by P-Solubilizing Microorganisms in Maize. Microorganisms 2019, 7, 38. [Google Scholar] [CrossRef] [Green Version]
- Kotan, R.; Sahin, F.; Demirci, E.; Ozbek, A.; Eken, C.; Miller, S.A. Evaluation of Antagonistic Bacteria for Biological Control of Fusar- Iumdry Rot of Potato. Phytopathology 1999, 89, 41. [Google Scholar]
- Eşitken, A.; Karlidaǧ, H.; Ercişli, S.; Şahin, F. Effects of Foliar Application of Bacillus Subtilis OSU-142 on the Yield, Growth and Control of Shot-Hole Disease (Coryneum Blight) of Apricot. Gartenbauwissenschaft 2002, 67, S139–S142. [Google Scholar]
- Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Chiarini, L. Efficacy of Burkholderia Cepacia MCI 7 in Disease Suppression and Growth Promotion of Maize. Biol. Fertil. Soils 2000, 31, 225–231. [Google Scholar] [CrossRef]
- Ciccillo, F.; Fiore, A.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Chiarini, L. Effects of Two Different Application Methods of Burkholderia Ambifaria MCI 7 on Plant Growth and Rhizospheric Bacterial Diversity. Environ. Microbiol. 2002, 4, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.; Bevivino, A.; Tabacchioni, S.; Dalmastri, C. Inoculation of Burkholderia Cepacia, Pseudomonas Fluorescens and Enterobacter Sp. on Sorghum Bicolor: Root Colonization and Plant Growth Promotion of Dual Strain Inocula. Soil Biol. Biochem. 1998, 30, 81–87. [Google Scholar] [CrossRef]
- Nielsen, M.N.; Sørensen, J.; Fels, J.; Pedersen, H.C. Secondary Metabolite- and Endochitinase-Dependent Antagonism toward Plant-Pathogenic Microfungi of Pseudomonas Fluorescens Isolates from Sugar Beet Rhizosphere. Appl. Environ. Microbiol. 1998, 64, 3563–3569. [Google Scholar] [CrossRef] [Green Version]
- Volpe, V.; Chitarra, W.; Cascone, P.; Volpe, M.G.; Bartolini, P.; Moneti, G.; Pieraccini, G.; Di Serio, C.; Maserti, B.; Guerrieri, E.; et al. The Association With Two Different Arbuscular Mycorrhizal Fungi Differently Affects Water Stress Tolerance in Tomato. Front. Plant Sci. 2018, 9, 1480. [Google Scholar] [CrossRef] [PubMed]
- Fracasso, A.; Telò, L.; Lanfranco, L.; Bonfante, P.; Amaducci, S. Physiological Beneficial Effect of Rhizophagus Intraradices Inoculation on Tomato Plant Yield under Water Deficit Conditions. Agronomy 2020, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Matarese, F.; Sarrocco, S.; Gruber, S.; Seidl-Seiboth, V.; Vannacci, G. Biocontrol of Fusarium Head Blight: Interactions between Trichoderma and Mycotoxigenic Fusarium. Microbiology 2012, 158, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Vinci, G.; Cozzolino, V.; Mazzei, P.; Monda, H.; Spaccini, R.; Piccolo, A. An Alternative to Mineral Phosphorus Fertilizers: The Combined Effects of Trichoderma Harzianum and Compost on Zea Mays, as Revealed by 1 H NMR and GC-MS Metabolomics. PLoS ONE 2018, 13, e0209664. [Google Scholar]
- Fiorini, L.; Guglielminetti, L.; Mariotti, L.; Curadi, M.; Picciarelli, P.; Scartazza, A.; Sarrocco, S.; Vannacci, G. Trichoderma Harzianum T6776 Modulates a Complex Metabolic Network to Stimulate Tomato Cv. Micro-Tom Growth. Plant Soil 2016, 400, 351–366. [Google Scholar] [CrossRef]
- Middelhoven, W.J.; Muylwijk, C.M. High-Affinity Amylolytic Enzymes Produced by the Mould Trichoderma Harzianum. Appl. Microbiol. Biotechnol. 1986, 23, 400–403. [Google Scholar] [CrossRef]
- Malusá, E.; Sas-Paszt, L.; Ciesielska, J. Technologies for Beneficial Microorganisms Inocula Used as Biofertilizers. Sci. World J. 2012, 2012, 491206. [Google Scholar] [CrossRef] [PubMed]
- Lutts, S.; Benincasa, P.; Wojtyla, L.; Kubala, S.; Pace, R.; Lechowska, K.; Quinet, M.; Garnczarska, M. Seed Priming: New Comprehensive Approaches for an Old Empirical Technique. In New Challenges in Seed Biology-Basic and Translational Research Driving Seed Technology; Araujo, S., Balestrazzi, A., Eds.; IntechOpen: London, UK, 2016. [Google Scholar]
- Boari, A.; Zuccari, D.; Vurro, M. “Microbigation”: Delivery of Biological Control Agents through Drip Irrigation Systems. Irrig. Sci. 2008, 26, 101–107. [Google Scholar] [CrossRef]
- Swaminathan, J.; van Koten, C.; Henderson, H.V.; Jackson, T.A.; Wilson, M.J. Formulations for Delivering Trichoderma Atroviridae Spores as Seed Coatings, Effects of Temperature and Relative Humidity on Storage Stability. J. Appl. Microbiol. 2016, 120, 425–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slater, S.C.; Goldman, B.S.; Goodner, B.; Setubal, J.C.; Farrand, S.K.; Nester, E.W.; Burr, T.J.; Banta, L.; Dickerman, A.W.; Paulsen, I.; et al. Genome Sequences of Three Agrobacterium Biovars Help Elucidate the Evolution of Multichromosome Genomes in Bacteria. J. Bacteriol. 2009, 191, 2501–2511. [Google Scholar] [CrossRef] [Green Version]
- Wisniewski-Dyé, F.; Borziak, K.; Khalsa-Moyers, G.; Alexandre, G.; Sukharnikov, L.O.; Wuichet, K.; Hurst, G.B.; McDonald, W.H.; Robertson, J.S.; Barbe, V.; et al. Azospirillum Genomes Reveal Transition of Bacteria from Aquatic to Terrestrial Environments. PLoS Genet. 2011, 7, e1002430. [Google Scholar] [CrossRef] [PubMed]
- Robson, R.L.; Jones, R.; Robson, R.M.; Schwartz, A.; Richardson, T.H. Azotobacter Genomes: The Genome of Azotobacter Chroococcum NCIMB 8003 (ATCC 4412). PLoS ONE 2015, 10, e0127997. [Google Scholar] [CrossRef] [Green Version]
- Noar, J.D.; Bruno-Bárcena, J.M. Complete Genome Sequences of Azotobacter Vinelandii Wild-Type Strain CA and Tungsten-Tolerant Mutant Strain CA6. Genome Announc. 2013, 1, e00313-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus Amyloliquefaciens Clades Associated with Strains DSM 7 T and FZB42 T: A Proposal for Bacillus Amyloliquefaciens Subsp. Amyloliquefaciens Subsp. Nov. and Bacillus Amyloliquefaciens Subsp. Plantarum Subsp. Nov. Based on Complete Gen. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative Analysis of the Complete Genome Sequence of the Plant Growth-Promoting Bacterium Bacillus Amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, M.W.; Ramaiya, P.; Nelson, B.A.; Brody-Karpin, S.D.; Zaretsky, E.J.; Tang, M.; Lopez de Leon, A.; Xiang, H.; Gusti, V.; Clausen, I.G.; et al. Complete Genome Sequence of the Industrial Bacterium Bacillus Licheniformis and Comparisons with Closely Related Bacillus Species. Genome Biol. 2004, 5, r77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, K.K.; Dey, R.; Sherathia, D.; Vanpariya, S.; Patel, I.; Dalsania, T.; Savsani, K.; Sukhadiya, B.; Mandaliya, M.; Thomas, M.; et al. Draft Genome Sequence of a Moderately Halophilic Bacillus Megaterium Strain, MSP20.1, Isolated from a Saltern of the Little Rann of Kutch, India. Genome Announc. 2014, 2, e01134-13. [Google Scholar] [CrossRef] [Green Version]
- Barbe, V.; Cruveiller, S.; Kunst, F.; Lenoble, P.; Meurice, G.; Sekowska, A.; Vallenet, D.; Wang, T.; Moszer, I.; Médigue, C.; et al. From a Consortium Sequence to a Unified Sequence: The Bacillus Subtilis 168 Reference Genome a Decade Later. Microbiology 2009, 155, 1758–1775. [Google Scholar] [CrossRef] [Green Version]
- Kuramae, E.E.; Derksen, S.; Schlemper, T.R.; Dimitrov, M.R.; Costa, O.Y.A.; da Silveira, A.P.D. Sorghum Growth Promotion by Paraburkholderia Tropica and Herbaspirillum Frisingense: Putative Mechanisms Revealed by Genomics and Metagenomics. Microorganisms 2020, 8, 725. [Google Scholar] [CrossRef]
- Miguel, R.N.; Barret, M.; Morrisey, J.P.; Germaine, K.; Francisco, M.G.; Barahona, E.; Navazo, A.; María, S.C.; Moynihan, J.A.; Giddens, S.R.; et al. Genome Sequence of the Biocontrol Strain Pseudomonas Fluorescens F113. J. Bacteriol. 2012, 194, 1273–1274. [Google Scholar]
- Nelson, K.E.; Weinel, C.; Paulsen, I.T.; Dodson, R.J.; Hilbert, H.; Martins dos Santos, V.A.P.; Fouts, D.E.; Gill, S.R.; Pop, M.; Holmes, M.; et al. Complete Genome Sequence and Comparative Analysis of the Metabolically Versatile Pseudomonas Putida KT2440. Environ. Microbiol. 2002, 4, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Jiao, Z.; Li, L.; Wu, D.; Crowley, D.E.; Wang, Y.; Wu, W. Draft Genome Sequence of Rahnella Aquatilis Strain HX2, a Plant Growth-Promoting Rhizobacterium Isolated from Vineyard Soil in Beijing, China. J. Bacteriol. 2012, 194, 6646–6647. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Paredes, A.; Valdés, G.; Nuti, M. Ecosystem Functions of Microbial Consortia in Sustainable Agriculture. Agronomy 2020, 10, 1902. [Google Scholar] [CrossRef]
- Stamenković, S.; Beškoski, V.; Karabegović, I.; Lazić, M.; Nikolić, N. Microbial Fertilizers: A Comprehensive Review of Current Findings and Future Perspectives. Span. J. Agric. Res. 2018, 16, e09R01. [Google Scholar] [CrossRef] [Green Version]
- Thomloudi, E.-E.; Tsalgatidou, P.C.; Douka, D.; Spantidos, T.-N.; Dimou, M.; Venieraki, A.; Katinakis, P. Multistrain versus Single-Strain Plant Growth Promoting Microbial Inoculants-The Compatibility Issue. Hell. Plant Prot. J. 2019, 12, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Che, S.; Men, Y. Synthetic Microbial Consortia for Biosynthesis and Biodegradation: Promises and Challenges. J. Ind. Microbiol. Biotechnol. 2019, 46, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Armanhi, J.S.L.; de Souza, R.S.C.; de Brito Damasceno, N.; de Araújo, L.M.; Imperial, J.; Arruda, P. A Community-Based Culture Collection for Targeting Novel Plant Growth-Promoting Bacteria from the Sugarcane Microbiome. Front. Plant Sci. 2018, 8, 2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [Green Version]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The “Secrets” of a Multitalented Biocontrol Agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Li, Y.; Yu, C.; Wang, Q.Q.; Wang, M.; Sun, J.; Gao, J.X.; Chen, J. Effect of Trichoderma Harzianum on Maize Rhizosphere Microbiome and Biocontrol of Fusarium Stalk Rot. Sci. Rep. 2017, 7, 1771. [Google Scholar] [CrossRef]
- Thonar, C.; Lekfeldt, J.D.S.; Cozzolino, V.; Kundel, D.; Kulhánek, M.; Mosimann, C.; Neumann, G.; Piccolo, A.; Rex, M.; Symanczik, S.; et al. Potential of Three Microbial Bio-Effectors to Promote Maize Growth and Nutrient Acquisition from Alternative Phosphorous Fertilizers in Contrasting Soils. Chem. Biol. Technol. Agric. 2017, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Shukla, N.; Awasthi, R.P.; Rawat, L.; Kumar, J. Biochemical and Physiological Responses of Rice (Oryza Sativa, L.) as Influenced by Trichoderma Harzianum under Drought Stress. Plant Physiol. Biochem. 2012, 54, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Mullins, A.J.; Murray, J.A.H.; Bull, M.J.; Jenner, M.; Jones, C.; Webster, G.; Green, A.E.; Neill, D.R.; Connor, T.R.; Parkhill, J.; et al. Genome Mining Identifies Cepacin as a Plant-Protective Metabolite of the Biopesticidal Bacterium Burkholderia Ambifaria. Nat. Microbiol. 2019, 4, 996–1005. [Google Scholar] [CrossRef]
- Bernabeu, P.R.; García, S.S.; López, A.C.; Vio, S.A.; Carrasco, N.; Boiardi, J.L.; Luna, M.F. Assessment of Bacterial Inoculant Formulated with Paraburkholderia Tropica to Enhance Wheat Productivity. World J. Microbiol. Biotechnol. 2018, 34, 81. [Google Scholar] [CrossRef]
- García, S.S.; Bernabeu, P.R.; Vio, S.A.; Cattelan, N.; García, J.E.; Puente, M.L.; Galar, M.L.; Prieto, C.I.; Luna, M.F. Paraburkholderia Tropica as a Plant-Growt-promoting Bacterium in Barley: Characterization of Tissues Colonization by Culture-Dependent and -Independent Techniques for Use as an Agronomic Bioinput. Plant Soil 2019, 451, 106–189. [Google Scholar]
- Schoebitz, M.; López, M.D.; Serri, H.; Aravena, V.; Zagal, E.; Roldán, A. Characterization of Bioactive Compounds in Blueberry and Their Impact on Soil Properties in Response to Plant Biostimulants. Commun. Soil Sci. Plant Anal. 2019, 50, 2482–2494. [Google Scholar] [CrossRef]
- Olivares, F.L.; Aguiar, N.O.; Rosa, R.C.C.; Canellas, L.P. Substrate Biofortification in Combination with Foliar Sprays of Plant Growth Promoting Bacteria and Humic Substances Boosts Production of Organic Tomatoes. Sci. Hortic. (Amsterdam) 2015, 183, 100–108. [Google Scholar] [CrossRef]





| PGPMs | Strain | Origin | Country | Properties | References |
|---|---|---|---|---|---|
| Acaulospora morrowiae | CL290 | Rhizosphere | USA | PGP | [34] |
| Agrobacterium radiobacter * | AR 39 | Soil near peach tree | Italy | Biocontrol/PGP | Unpublished results |
| Azospirillum brasilense * | ATTC 29710 | Cynodon dactylon rhizosphere | USA | N-fixation | [35] |
| Azospirillum brasilense * | NCCB 78036 | Soil under soy field | India | N-fixation | Unpublished results |
| Azospirillum lipoferum | CRT1 | Field grown maize | France | N-fixation | [36] |
| Azotobacter chroococcum | 76A | Soil | Italy | N-fixation | [37] |
| Azotobacter chroococcum * | DSM 2286 | Unknown | unknown | N-fixation | [38,39] |
| Azotobacter chroococcum * | LS132 | Rhizosphere | Italy | N-fixation | Unpublished results |
| Azotobacter chroococcum * | LS163 | Rhizosphere | Italy | N-fixation | Unpublished results |
| Azotobacter chroococcum | S-5 | Unknown | Iran | N-fixation | [38,39] |
| Azotobacter vinelandii * | DSM 2289 | Unknown | unknown | Siderophore production, N-fixation | [40,41] |
| Bacillus sp. | BV84 | Grape leafs | Italy | Biocontrol/PGP | Unpublished results |
| Bacillus amyloliquefaciens * | BA41 | Wheat rhizosphere | Italy | Biocontrol/PGP | Unpublished results |
| Bacillus amyloliquefaciens | FZB42 | Plant pathogen infested soil | Germany | Biocontrol/PGP | [42] |
| Bacillus amyloliquefaciens * | LMG 9814 | Soil | UK | Alpha-amylase, alpha-glucosidase, iso-amylase production | Unpublished results |
| Bacillus atrophaeus | ABI02A | NA | Germany | PGP | [43] |
| Bacillus licheniformis * | PS141 | Rhizosphere | Italy | Indole acetic acid (IAA) production | Unpublished results |
| Bacillus megaterium | M3 | Rice | unknown | P-solubilization | [44,45] |
| Bacillus megaterium | PMC 1855 | Unknown | unknown | P-solubilization | [46] |
| Bacillus pumilus * | LMG 24415 | Soil | Ecuador | PGP | [47] |
| Bacillus simple | R49538 | Unknown | Ecuador | PGP/IAA production | [47] |
| Bacillus subtilis | FZB24 WG | NA | Germany | Biocontrol/PGP | [48,49] |
| Bacillus subtilis * | LMG 23370 | Forest soil | India | Biocontrol/PGP | Unpublished results |
| Bacillus subtilis * | LMG 24418 | Soil | Ecuador | PGP | [47] |
| Bacillus subtilis | OSU-142 | pepper | unknown | N-fixation, biocontrol | [50,51] |
| Burkholderia ambifaria * | MCI 7 | Maize rhizosphere | Italy | PGP | [52,53] |
| Burkholderia ambifaria * | PHP7/LMG 11351 | Maize rhizosphere | France | PGP | [54] |
| Gigaspora gigantea | PA125 | Rhizosphere | USA | PGP | [34] |
| Gigaspora rosea | NY328A | Rhizosphere | USA | PGP | [34] |
| Komagataella pastoris * | PP59 | Grape rhizosphere | Italy | PGP | Unpublished results |
| Paenibacillus sp | R47065 | Unknown | Ecuador | PGP/IAA production | [47] |
| Paraburkholderia tropica | MDIIIAzo225 | Maize rhizosphere | Italy | N-fixation | Unpublished results |
| Pseudomonas granadensis ** | A23/T3c | Soil | Italy | PGP | [54] |
| Pseudomonas fluorescens * | DR54 | Sugar beet rhizosphere | Denmark | Biocontrol | [55] |
| Pseudomonas putid | P1-20/08 | Soil | Ecuador | PGP | [47] |
| Pseudomonas sp. * | PN53 | Grass rhizosphere | Italy | PGP | Unpublished results |
| Rahnella aquatilis ** | BB23/T4d | soil | Italy | PGP | [54] |
| Raoultella terrigena * | FS152 | Rhizosphere | Italy | Phytase activity, siderophore production | Unpublished results |
| Rhizophagus intraradices§ | FR121 § | - | - | Tolerance to abiotic /biotic stress | [56,57] |
| Septoglomus constrictum | FL328 | Rhizosphere | USA | PGP | Unpublished results |
| Streptomyces sp. | SA 51 | Rhizosphere | Italy | Biocontrol | Unpublished results |
| Trichoderma gamsii | 6085 | uncultivated soil | Crimea (UA) | Biocontrol | [58] |
| Trichoderma harzianum | OmG-08 | Orchid roots | Germany | P-solubilization | [59] |
| Trichoderma harzianum | OmG-16 | NA | Germany | P-solubilization | [49] |
| Trichoderma harzianum | T6776 | Soil | Italy | Biocontrol/PGP | [60] |
| Trichoderma harzianum * | TH01 | Grass soil and rhizosphere | Italy | PGP | Unpublished results |
| Trichoderma harzianum * | CBS 354.33/ ATCC 48131 | Soil | USA | Chitinase production, biocontrol | [61] |
| PGP Microbial Species | Representative Strain | GenBank Accession No. | RefSeq Accession No. | Reference |
|---|---|---|---|---|
| Agrobacterium radiobacter | K84 | chromosome 1/2: CP000628.1/CP000629.1 | chromosome 1/2: NC_011985.1/NC_011983.1 | [66] |
| Azospirillum brasilense | Sp7 | CP012914.1 | NZ_CP012914.1 | |
| Azospirillum lipoferum | 4B | FXBR00000000.1 | NZ_FXBR00000000.1 | [67] |
| Azotobacter chroococcum | NCIMB 8003 | CP010415.1 | NZ_CP010415.1 | [68] |
| Azotobacter chroococcum | DSM 2286 | SRX5354579 | ||
| Azotobacter vinelandii | CA | CP005094.1 | NC_021149.1 | [69] |
| Bacillus amyloliquefaciens | DSM 7 | FN597644.1 | NC_014551.1 | [70] |
| Bacillus amyloliquefaciens subsp. plantarum; now Bacillus velezensis | FZB42 | CP000560.1 | [71] | |
| Bacillus atrophaeus subsp. globigii | SRCM101359 | CP021500.1 | NZ_CP021500.1 | |
| Bacillus licheniformis | DSM 13, ATCC 14580 | CP000002.3 | NC_006270.3 | [72] |
| Bacillus megaterium | MSP20.1 | CP009920.1 | NZ_CP009920.1 | [73] |
| Bacillus pumilus | SH-B9 | CP011007.1 | NZ_CP011007.1 | |
| Bacillus subtilis subsp. subtilis | 168 | AL009126.3 | NC_000964.3 | [74] |
| Bacillus simplex | SH-B26 | CP011008.1 | NZ_CP011008.1 | |
| Burkholderia ambifaria | MC40-6 | chromosome 1, 2, 3: CP001025.1, CP001026.1, CP001027.1 | chromosome 1, 2, 3: NC_010551.1, NC_010552.1, NC_010557.1 | |
| Komagataella pastoris (Pichia pastoris) | ATCC 28485 | chromosome 1, 2, 3, 4: CP014584.1, CP014585.1, CP014586.1, CP014587.1 | ||
| Paraburkholderia tropica | IAC135 | chromosome A, B, C, D, E: CP049134.1, CP049135.1, CP049136.1, CP049137.1, CP049138.1 | chromosome A, B, C, D, E: NZ_CP049134.1, NZ_CP049135.1, NZ_CP049136.1, NZ_CP049137.1, NZ_CP049138.1 | [75] |
| Pseudomonas fluorescens | F113 | CP003150.1 | NC_016830.1 | [76] |
| Pseudomonas granadensis | LMG 27940 | chromosome I: LT629778.1 | NZ_LT629778.1 | |
| Pseudomonas putida | KT2440 | AE015451.2 | NC_002947.4 | [77] |
| Rahnella aquatilis | HX2 | chromosome, plasmids PRA1 and PRA2 & PRA22: CP003403.1, CP003404.1, CP003405.1, CP003406.1 | NC_017047.1, NC_017060.1, NC_017807.1, NC_017773.1 | [78] |
| Raoultella terrigena | NCTC13098 | LR131271.1 | NZ_LR131271.1 | |
| Trichoderma harzianum | CBS 226.95 | GCA_003025095.1 | GCF_003025095.1 |
| Strain | A. radiobacter AR39 | A. brasilense ATCC 29710 | A. brasilense NCCB 78036 | A. chroococcum DSM 2286 | A. chroococcum LS132 | A. chroococcum LS136 | A. vinelandii DSM 2289 | Bacillus sp. BV84 | B. amyloliquefaciens BA41 | B. amyloliquefaciens LMG 9814 | B. licheniformis PS141 | B. pumilus LMG 24415 | B. subtilis LMG 23370 | B. subtilis LMG 24418 | B. ambifaria LMG 11351 | B. ambifaria MCI 7 | B. ambifaria LMG 11351 | K. pastoris PP59 | P. tropica MDIIIAzo225 | Pseudomonas sp. PN53 | P. fluorescens DR54 | P. granadensis A23/T3c | R. aquatilis BB23/T4d | R. terrigena FS152 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. radiobacter AR39 | ||||||||||||||||||||||||
| A. brasilense ATCC 29710 | + | |||||||||||||||||||||||
| A. brasilense NCCB 78036 | + | + | ||||||||||||||||||||||
| A. chroococcum DSM 2286 | - | - | - | |||||||||||||||||||||
| A. chroococcum LS132 | + | + | + | - | ||||||||||||||||||||
| A. chroococcum LS136 | + | + | + | - | + | |||||||||||||||||||
| A. vinelandii DSM 2289 | + | + | + | - | + | + | ||||||||||||||||||
| Bacillus sp. BV84 | + | - | + | + | + | + | + | |||||||||||||||||
| B. amyloliquefaciens BA41 | + | - | + | + | + | + | + | + | ||||||||||||||||
| B. amyloliquefaciens LMG 9814 | + | nc | + | - | + | + | + | + | + | |||||||||||||||
| B. licheniformis PS141 | + | + | + | - | + | + | + | - | - | - | ||||||||||||||
| B. pumilus LMG 24415 | + | + | + | + | + | + | + | - | - | - | - | |||||||||||||
| B. subtilis LMG 23370 | + | + | + | + | + | + | + | - | - | - | + | - | ||||||||||||
| B. subtilis LMG 24418 | + | + | + | + | + | + | + | - | - | - | + | - | - | |||||||||||
| B. ambifaria LMG 11351 | + | + | + | - | + | + | + | + | + | + | + | + | + | - | ||||||||||
| B. ambifaria MCI 7 | + | - | + | - | + | + | + | + | + | + | + | + | + | + | + | |||||||||
| K. pastoris PP59 | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + | nc | + | |||||||
| P. tropica MDIIIAzo225 | + | + | + | - | + | + | + | + | + | nc | + | + | nc | + | + | + | + | + | ||||||
| Pseudomonas sp. PN53 | + | + | + | + | + | + | - | - | + | + | + | + | + | + | + | + | + | + | + | |||||
| P. fluorescens DR54 | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | nc | ||||
| P. granadensis A23/T3c | + | + | + | - | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |||
| R. aquatilis BB23/T4d | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| R. terrigena FS152 | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Bacteria | T. harzianum ATCC 48131 | T. harzianum TH01 |
|---|---|---|
| Azotobacter brasilense ATCC 29710 | nc | + |
| Azospirillum brasilense NCCB 78036 | - | + |
| Azotobacter chroococcum DSM 2286 | - | nc |
| Azotobacter chroococcum LS132 | + | + |
| Agrobacterium radiobacter AR39 | - | - |
| Azotobacter chroococcum LS163 | - | + |
| Azotobacter vinelandii DSM 2289 | + | + |
| Bacillus sp. BV84 | - | + |
| Bacillus amyloliquefaciens BA41 | - | - |
| Bacillus amyloliquefaciens LMG 9814 | - | - |
| Bacillus licheniformis PS141 | + | + |
| Bacillus pumilus LMG 24415 | - | - |
| Bacillus subtilis LMG 23370 | - | - |
| Bacillus subtilis LMG 24418 | - | - |
| Burkholderia ambifaria LMG 11351 | - | - |
| Burkholderia ambifaria MCI 7 | - | - |
| Komagataella pastoris PP59 | + | + |
| Paraburkholderia tropica MDIIIAzo225 | + | nc |
| Pseudomonas sp. PN53 | - | + |
| Pseudomonas granadensis A23/T3c | + | + |
| Pseudomonas fluorescens DR54 | + | nc |
| Ranhella aquatilis BB23/T4d | - | + |
| Raoultella terrigena FS152 | - | + |
| Strain | BS1 | BS2 | BS3 | BS4 |
|---|---|---|---|---|
| Azotobacter chroococcum LS132 | + | + | - | nc |
| Azotobacter vinelandii DSM 2289 | + | + | - | nc |
| Bacillus sp. BV84 | + | + | - | nc |
| Bacillus amyloliquefaciens LMG 9814 | + | + | - | nc |
| Bacillus licheniformis PS141 | - | + | - | nc |
| Burkholderia ambifaria MCI 7 | + | + | - | nc |
| Paraburkholderia tropica MDIIIAzo225 | - | + | - | nc |
| Pichia pastoris PP59 | - | + | - | nc |
| Pseudomonas sp. A23/T3c | + | + | - | nc |
| Pseudomonas fluorescens DR54 | + | + | - | nc |
| Rahnella aquatilis BB23/T4d | - | + | - | nc |
| Trichoderma harzianum TH01 | + | + | + | nc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabacchioni, S.; Passato, S.; Ambrosino, P.; Huang, L.; Caldara, M.; Cantale, C.; Hett, J.; Del Fiore, A.; Fiore, A.; Schlüter, A.; et al. Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture. Microorganisms 2021, 9, 426. https://doi.org/10.3390/microorganisms9020426
Tabacchioni S, Passato S, Ambrosino P, Huang L, Caldara M, Cantale C, Hett J, Del Fiore A, Fiore A, Schlüter A, et al. Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture. Microorganisms. 2021; 9(2):426. https://doi.org/10.3390/microorganisms9020426
Chicago/Turabian StyleTabacchioni, Silvia, Stefania Passato, Patrizia Ambrosino, Liren Huang, Marina Caldara, Cristina Cantale, Jonas Hett, Antonella Del Fiore, Alessia Fiore, Andreas Schlüter, and et al. 2021. "Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture" Microorganisms 9, no. 2: 426. https://doi.org/10.3390/microorganisms9020426
APA StyleTabacchioni, S., Passato, S., Ambrosino, P., Huang, L., Caldara, M., Cantale, C., Hett, J., Del Fiore, A., Fiore, A., Schlüter, A., Sczyrba, A., Maestri, E., Marmiroli, N., Neuhoff, D., Nesme, J., Sørensen, S. J., Aprea, G., Nobili, C., Presenti, O., ... Bevivino, A. (2021). Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture. Microorganisms, 9(2), 426. https://doi.org/10.3390/microorganisms9020426