3.3. Determination of Gastrointestinal Survival and Biogenic Amine Formation
Survival in a gastrointestinal environment is a prerequisite for oral probiotic formulations to colonize the intestinal tract. Tolerance to artificial gastric juice and intestinal juice were tested among strains without hemolytic activity. A total of 18 out of 22 strains evaluated could survive through both gastric and intestinal juice (
Figure 1).
The active metabolism of amino acids, usually decarboxylation in microorganisms, might result in high levels of biogenic amines, which would have negative effects on vasoactivity and psychoactivity, or as the precursors of carcinogenic nitrosamines [
35]. Thus, biogenic amine formation is a very important evaluation index for the safety of probiotics. The biogenic amine formation by the isolates was determined using a screening assay for amino acid decarboxylation from ornithine and histidine. Ten isolates having strong biogenic amine formation were excluded through this assay. Representative results of strains without biogenic amine formation are displayed in
Table 2.
Thus far, eight strains were screened out after the primary safety assessment and gastrointestinal tolerance evaluation. These strains were retained for subsequent characterization of probiotic properties. Three of them were Lactobacillus (N15-22, X16-68, X16-69), similar to L. senioris and L. curieae. The other five strains were Bacillales, among which two were Bacillus, similar to B. licheniformis (N17-02) and B. ginsengihumi (X16-66); the left three were assigned as Bacillus megaterium (N17-12), Virgibacillus halophilus (Y11-38) and Paenibacillus chibensis (Y13-51). Sequences of the 16S rRNA gene from these isolates were deposited in the GenBank, National Center of Biotechnology Information (NCBI) database under accession No. MW578436-MW578443 (accessed on 12 February 2021).
Overall, all of these eight strains had reasonable chance of survival in the simulated gastrointestinal environment, while Y11-38 and Y13-51 showed the most prominent adaptability (
Table 3). Relatively higher survival rates were found in
Bacillus strains than in
Lactobacillus strains. The survival rates of
Bacillus strains were in the range of 67.18–96.89% and rates of
Lactobacillus strains were in the range of 3.60–25.00% after exposure to artificial gastric juice for 4 h. Similarly, in artificial intestinal juice they were 45.18–48.06% for
Bacillus strains and 27.36–43.48% for
Lactobacillus strains. This observation was consistent with previous reports about the greater viability of
Bacillus strains than
Lactobacillus strains when exposed to simulated gastrointestinal conditions or tea and coffee brewing [
16,
17,
36].
3.4. Auto-Aggregation and Cell Surface Hydrophobicity
Probiotic candidates should be able to attach onto the epithelium lining of the ileum in the host before they could provide health benefits, thus their adhesion ability was assessed through auto-aggregation and cell surface hydrophobicity assay. High auto-aggregation is correlated with effective adhesion and subsequent colonization in the gastrointestinal tract. The auto-aggregation of these eight strains increased with the incubation time, reaching a range of 26.12–94.97% at 6 h and 52.53–95.56% at 24 h, respectively (
Table 4). Among them, N15-22 and N17-02 had the highest auto-aggregation (95.56% and 93.69%) at 24 h. However, given that chyme spends on average 4–8 h in the ileum [
18],
B. licheniformis N17-02 was considered the best one due to the fact that its auto-aggregation rapidly increased in the first 6 h.
Hydrophobicity reveals the relative tendency of a substance to prefer nonaqueous rather than aqueous environments. A high cell surface hydrophobicity of a probiotic candidate indicates its probability to attach onto the epithelial lining of the intestine and resist the quick expelling from intestines. Here, the adhesion ability to hydrocarbons dichloromethane and ethyl-acetate were measured (
Table 4). Interestingly, the hydrophobicity measurements in three
Lactobacillus strains were higher than that of strains in Bacillales for both dichloromethane and ethyl-acetate, as high as 90.76–98.05% and 90.59–97.83%, respectively. Compared with the hydrophobicity reported previously, such as
Bacillus strains to ethyl-acetate [
21,
29] and
Lactobacillus isolates to dichloromethane [
37], the hydrophobicity measurements of the strains evaluated here were comparable or even higher.
In general, based on the auto-aggregation and hydrophobicity, these eight strains have good adhesion ability. Probiotics that aggregate and accumulate in sufficient quantities to form an immune barrier could protect the gut from pathogens, maintain the balance and stability of intestinal flora and perform the other functions of probiotics [
38].
3.5. Antibacterial Activity and Co-Aggregation
The antibacterial activity of isolates was tested against enteric bacterial pathogens, including
Staphylococcus aureus,
Salmonella paratyphi,
Escherichia coli and
Bacillus subtilis (
Table 5). For Bacillales strains, significant inhibition was observed only in N17-02 against
S. aureus and N17-12 against
S. paratyphi and
E. coli. The original supernatants of
Lactobacillus strains culture had significant inhibitory effects against most pathogens tested except N15-22 against
E. coli, but the antibacterial activity decreased after adjusting the supernatant to neutral. Thus, acid production might be an essential component of antibacterial activity in
Lactobacillus strains, or acidic conditions might be beneficial for the function of their bacteriostatic substances. Similarly, organic acids including lactic acid were reported to be responsible for the antagonistic effect of several lactic acid bacteria derived from fermented food [
39].
From the point of view of co-aggregation, these eight strains had co-aggregation at 46.67–69.18%, 50.98–67.49%, 48.00–59.36% and 70.78–82.10% to
S. aureus,
S. paratyphi,
E. coli and
B. subtilis, respectively (
Table 5). These data implied that these isolates had good chances to co-aggregate with pathogens to interfere with their retention and colonization in gastrointestinal tract. The highest co-aggregation with
S. aureus and
B. subtilis was observed in X16-69 and N17-02, respectively. The highest co-aggregation with
S. paratyphi and
E. coli was found in N17-12, together with its significant antibacterial activity against these two pathogens, thus
B. megaterium N17-12 was suggested as a good probiotic candidate from the point of view of the antibacterial potential. Three
B. megaterium strains were isolated recently from stingless bee honey collected across Malaysia and subjected to probiotic screening, but they were found to have no inhibition effect against six pathogenic bacteria, including
Salmonella thyphimurium and
E. coli [
40]. Thus, the probiotic property of
B. megaterium was strain specific.
3.6. Antibiotic Susceptibility
Given the possibility of transfer of antibiotic resistance genes from probiotic bacteria to gut pathogens, the antibiotic susceptibility was evaluated in these eight strains. According to their classification and mechanisms of action [
41], different antibiotics such as β-lactams (penicillin-G, ampicillin and cephalothin), macrolides (erythromycin), tetracyclines, aminoglycosides (gentamicin), glycopeptides (vancomycin) and chloramphenicol were chosen. Results indicated that each isolate was sensitive to at least three or more antibiotics (
Table 6), which verified their safe utilization. Synthetically, Y13-51 showed the highest vulnerability to antibiotics tested. Most Bacillales strains were resistant to penicillin-G, probably due to the narrow antibacterial spectrum of penicillin-G. The
Lactobacillus strains N15-22 and X16-68 were resistant to vancomycin and gentamicin, thus they might be applied to reduce the diarrhea caused by antibiotic damage to intestinal flora [
42]. It needs to be further evaluated whether these antibiotic resistances were transferable before application.
At this point, eight isolates had met the selection criteria, i.e., gastrointestinal tract stress tolerance, adhesion ability, antimicrobial activity and primary safety assessment, to be probiotic candidates. Then, the cholesterol-lowering capabilities and carbon source utilization were further investigated to explore their functional properties.
3.7. Cholesterol-Lowering Potential
Attention was first paid to the bile salt hydrolase (BSH), choloylglycine hydrolase (EC3.5.1.24), which is suggested to contribute to the detoxification of bile acid (BA), and is associated with the therapeutic effect of decreasing host cholesterol [
19,
43,
44]. Unfortunately, using TDCA and GDCA as BA substrates, the positive BSH activity as a white precipitation zone around the colonies was not observed in our experiment. Since non-concordance between BSH phenotypic assay and presence of
bsh gene was reported recently in
Enterococcus faecium isolated from rhizosphere [
30], we further searched for the
bsh gene in these eight isolates. According to the
bsh genes in similar strains as reference, touchdown PCR successfully retrieved six
bsh genes from five isolates, including N17-02, N17-12, Y11-38, X16-66 and N15-22. In particular, two different
bsh genes were identified in
L. senioris N15-22 (
Figure 2). High similarity was observed between the identified BSHs and the reference BSHs (
Figure 2,
Figure S2), which implied their functionality. A latest report in PNAS emphasized the substrate specificity of BSHs from
Lactobacillus, which governs bacterial fitness and host colonization [
45]. This might help explain the negative result in BSH activity assay, since only TDCA and GDCA were tested as substrates. Further analysis is needed to investigate whether these putative enzymes could act as functional BSHs.
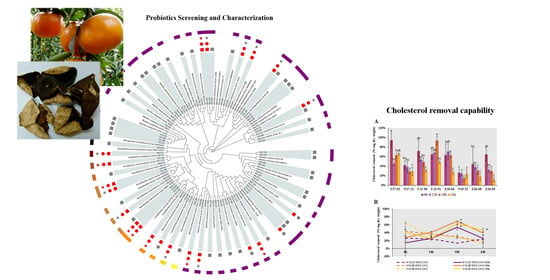
Then, the direct cholesterol removal capability, including cholesterol assimilation, absorption and adsorption, were evaluated in these eight strains. All of them displayed distinct cholesterol removal potential, and the removal rate ranged from 11.85% to 93.94% within 24 h (
Figure 3A). The highest cholesterol removal rates were found over 90% at 6 h in
B. licheniformis N17-02 and 18 h in
P. chibensis Y13-51, respectively. Compared with three
Lactobacillus strains, the removal rates were relatively higher in five isolates from Bacillales, ranging from 27.18% to 93.94%, which were higher than the removal rate 12.82–70.89% reported previously in
Bacillus strains isolated from fermented vegetables [
46]. To test whether bile salt could boost the relatively lower removal rate in
Lactobacillus strains N15-22, X16-68 and X16-69, GDCA and TDCA were added. Significant increase of removal rate was observed at 18 h in all three strains, to a maximum of 69.48% in
L. curieae X16-68 (
Figure 3B). The addition of bile salts might affect the cell surface structure to varying degrees, thus promoting cholesterol removal rate [
19].
Two possible mechanisms have been proposed regarding the hypocholesterolemia effect of probiotics: one is the direct binding or assimilation of cholesterol by probiotics and the other one is bile salt deconjugation catalyzed by BSH [
19]. Intestinal bacterial BSHs cleave glycine or taurine from conjugated BAs, an essential upstream step for the production of deconjugated and secondary BAs. Deconjugated BAs are less soluble and resorbable than the conjugated ones in the intestine, thus rendering them easier to be excreted in feces and subsequently more conjugated BA should be synthesized de novo from cholesterol, thereby reducing the cholesterol concentration [
19,
47]. Another consequence is the lower emulsifying ability of deconjugated bile salts compared to conjugated ones, which may result in lower lipid digestion and decreased absorption of cholesterol, fatty acids and monoglycerides in the intestine [
19,
44,
48]. The obvious cholesterol removal ability and the presence of
bsh genes in these isolates here suggested that they could be good probiotic candidates with hypocholesterolemia effect. Especially,
B. licheniformis N17-02 showed both remarkable cholesterol removal capability and presence of
bsh gene.
BSH was confirmed to be the key factor affecting the hypocholesterolemia activity of
Lactobacillus-fermented milk in hamsters [
43] and
Lactobacillus-yoghurt formulation in hypercholesterolemic adults [
49]. Nevertheless, over-extensive secondary BA might be associated with diseases such as steatosis and colorectal carcinoma [
19,
50,
51]. Therefore, more in vivo experiments are needed to support refined applications.
3.8. Carbohydrate Utilization Capability
The carbohydrate utilization profiles of these eight strains were investigated against two monosaccharides, three disaccharides, one trisaccharide, one tetrasaccharide and two sugar alcohols (
Table 7). All eight strains could make use of at least five carbon sources. In particular,
B. megaterium N17-12 showed significant growth on all the carbohydrate sources tested, including D-glucose, D-xylose, lactose, sucrose, cellobiose, stachyose, D-raffinose, sorbitol and mannitol. Such a wide carbohydrate utilization profile might suggest the growth advantage among GIT microbiota in host, where the competition in nutrition was fierce. Interestingly, N17-12 also displayed the strongest inhibitory effect against
S. paratyphi and E. coli (
Table 5), thus together confirming its great potential as probiotics. Other than being good as a probiotic to function in a host, the wide carbohydrate utilization profile also suggested its potential as a fermentation starter in fermented food or feed. For example, dough fermentation with
Lactobacillus was investigated recently to improve the nutritional quality of foodstuff [
52,
53]. The application of these eight strains in the fermentation of food or feed will be investigated in the future.