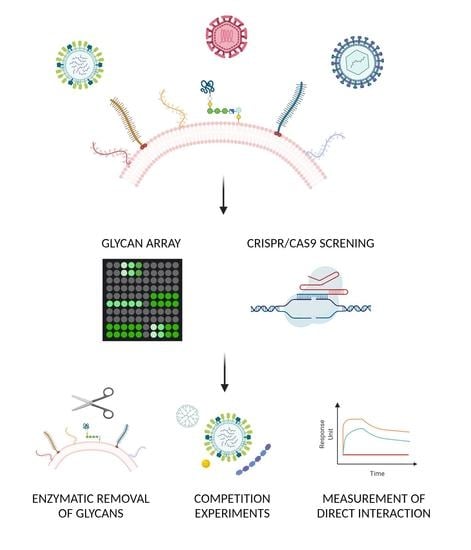
Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment
Abstract
:1. Introduction
2. Unbiased Methods: Unknown Carbohydrates
2.1. Glycomics
2.2. Glycan Arrays
2.3. Shotgun Glycomics
2.4. Genetic Approach
2.5. CHO Cells Mutagenized
2.6. Haploid Screening
2.7. CRISPR/Cas9 Libraries
3. Confirmatory Methods: Verification of the Glycan–Virus Interaction
3.1. Enzymatic Approach
3.2. Pharmacological Approach
3.3. Structural Approaches
4. Glycan–Virus Interactions
4.1. EV-D68 Dependency on Sialic Acid
4.2. SARS-CoV-2 Dependency on Glycans
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cagno, V.; Tseligka, E.D.; Jones, S.T.; Tapparel, C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses 2019, 11, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milewska, A.; Zarebski, M.; Nowak, P.; Stozek, K.; Potempa, J.; Pyrc, K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014, 88, 13221–13230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE 2011, 6, e23710. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057. [Google Scholar] [CrossRef]
- Lee, E.; Lobigs, M. E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. J. Virol. 2008, 82, 6024–6033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Varki, A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 2007, 446, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Wang, B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv. Nutr. 2012, 3, 465S–472S. [Google Scholar] [CrossRef] [Green Version]
- Wasik, B.R.; Barnard, K.N.; Parrish, C.R. Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol. 2016, 24, 991–1001. [Google Scholar] [CrossRef]
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; van Dieren, B.; Lang, Y.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, P.R.; Vinje, J.; Moe, C.L.; Baric, R.S. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 2004, 78, 3035–3045. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.C.; Arthur, C.M.; Wang, J.; Verkerke, H.; Josephson, C.D.; Kalman, D.; Roback, J.D.; Cummings, R.D.; Stowell, S.R. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 2021, 5, 1305–1309. [Google Scholar] [CrossRef]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [Green Version]
- Mende, M.; Bordoni, V.; Tsouka, A.; Loeffler, F.F.; Delbianco, M.; Seeberger, P.H. Multivalent glycan arrays. Faraday Discuss. 2019, 219, 9–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walther, T.; Karamanska, R.; Chan, R.W.; Chan, M.C.; Jia, N.; Air, G.; Hopton, C.; Wong, M.P.; Dell, A.; Malik Peiris, J.S.; et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013, 9, e1003223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amonsen, M.; Smith, D.F.; Cummings, R.D.; Air, G.M. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with alpha2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 2007, 81, 8341–8345. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, E.C.; Storm, R.J.; Bauer, J.; Johansson, S.M.; Lookene, A.; Angstrom, J.; Hedenstrom, M.; Eriksson, T.L.; Frangsmyr, L.; Rinaldi, S.; et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat. Med. 2011, 17, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Tuve, S.; Wang, H.; Jacobs, J.D.; Yumul, R.C.; Smith, D.F.; Lieber, A. Role of cellular heparan sulfate proteoglycans in infection of human adenovirus serotype 3 and 35. PLoS Pathog. 2008, 4, e1000189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haab, B.B.; Klamer, Z. Advances in Tools to Determine the Glycan-Binding Specificities of Lectins and Antibodies. Mol. Cell. Proteomics 2020, 19, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Geissner, A.; Reinhardt, A.; Rademacher, C.; Johannssen, T.; Monteiro, J.; Lepenies, B.; Thepaut, M.; Fieschi, F.; Mrazkova, J.; Wimmerova, M.; et al. Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci. USA 2019, 116, 1958–1967. [Google Scholar] [CrossRef] [Green Version]
- Byrd-Leotis, L.; Liu, R.; Bradley, K.C.; Lasanajak, Y.; Cummings, S.F.; Song, X.; Heimburg-Molinaro, J.; Galloway, S.E.; Culhane, M.R.; Smith, D.F.; et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc. Natl. Acad. Sci. USA 2014, 111, E2241–E2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Lasanajak, Y.; Song, X.; Hu, L.; Ramani, S.; Mickum, M.L.; Ashline, D.J.; Prasad, B.V.; Estes, M.K.; Reinhold, V.N.; et al. Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Mol. Cell. Proteomics 2014, 13, 2944–2960. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Mishra, S.; Song, X.; Lasanajak, Y.; Bradley, K.C.; Tappert, M.M.; Air, G.M.; Steinhauer, D.A.; Halder, S.; Cotmore, S.; et al. Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. J. Biol. Chem. 2012, 287, 44784–44799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carette, J.E.; Guimaraes, C.P.; Varadarajan, M.; Park, A.S.; Wuethrich, I.; Godarova, A.; Kotecki, M.; Cochran, B.H.; Spooner, E.; Ploegh, H.L.; et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 2009, 326, 1231–1235. [Google Scholar] [CrossRef] [Green Version]
- Jae, L.T.; Raaben, M.; Riemersma, M.; van Beusekom, E.; Blomen, V.A.; Velds, A.; Kerkhoven, R.M.; Carette, J.E.; Topaloglu, H.; Meinecke, P.; et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science 2013, 340, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Luteijn, R.D.; van Diemen, F.; Blomen, V.A.; Boer, I.G.J.; Manikam Sadasivam, S.; van Kuppevelt, T.H.; Drexler, I.; Brummelkamp, T.R.; Lebbink, R.J.; Wiertz, E.J. A Genome-Wide Haploid Genetic Screen Identifies Heparan Sulfate-Associated Genes and the Macropinocytosis Modulator TMED10 as Factors Supporting Vaccinia Virus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Riblett, A.M.; Blomen, V.A.; Jae, L.T.; Altamura, L.A.; Doms, R.W.; Brummelkamp, T.R.; Wojcechowskyj, J.A. A Haploid Genetic Screen Identifies Heparan Sulfate Proteoglycans Supporting Rift Valley Fever Virus Infection. J. Virol. 2016, 90, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Baggen, J.; Thibaut, H.J.; Staring, J.; Jae, L.T.; Liu, Y.; Guo, H.; Slager, J.J.; de Bruin, J.W.; van Vliet, A.L.; Blomen, V.A.; et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. USA 2016, 113, 1399–1404. [Google Scholar] [CrossRef] [Green Version]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An essential receptor for adeno-associated virus infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.L.; Eddy, R.L.; Shows, T.B.; Lau, J.T. Three genes that encode human beta-galactoside alpha 2,3-sialyltransferases. Structural analysis and chromosomal mapping studies. Glycobiology 1995, 5, 319–325. [Google Scholar] [CrossRef]
- Tanaka, A.; Tumkosit, U.; Nakamura, S.; Motooka, D.; Kishishita, N.; Priengprom, T.; Sa-Ngasang, A.; Kinoshita, T.; Takeda, N.; Maeda, Y. Genome-Wide Screening Uncovers the Significance of N-Sulfation of Heparan Sulfate as a Host Cell Factor for Chikungunya Virus Infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Baggen, J.; Thibaut, H.J.; Hurdiss, D.L.; Wahedi, M.; Marceau, C.D.; van Vliet, A.L.W.; Carette, J.E.; van Kuppeveld, F.J.M. Identification of the Cell-Surface Protease ADAM9 as an Entry Factor for Encephalomyocarditis Virus. MBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Baggen, J.; Liu, Y.; Lyoo, H.; van Vliet, A.L.W.; Wahedi, M.; de Bruin, J.W.; Roberts, R.W.; Overduin, P.; Meijer, A.; Rossmann, M.G.; et al. Bypassing pan-enterovirus host factor PLA2G16. Nat. Commun. 2019, 10, 3171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.S.L.; Yusa, K. Genome-wide CRISPR-Cas9 screening in mammalian cells. Methods 2019, 164-165, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; ten Oever, B.; Manicassamy, B. Genome-wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Muffat, J.; Omer Javed, A.; Keys, H.R.; Lungjangwa, T.; Bosch, I.; Khan, M.; Virgilio, M.C.; Gehrke, L.; Sabatini, D.M.; et al. Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc. Natl. Acad. Sci. USA 2019, 116, 9527–9532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petitjean, O.; Girardi, E.; Ngondo, R.P.; Lupashin, V.; Pfeffer, S. Genome-Wide CRISPR-Cas9 Screen Reveals the Importance of the Heparan Sulfate Pathway and the Conserved Oligomeric Golgi Complex for Synthetic Double-Stranded RNA Uptake and Sindbis Virus Infection. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Thamamongood, T.; Aebischer, A.; Wagner, V.; Chang, M.W.; Elling, R.; Benner, C.; García-Sastre, A.; Kochs, G.; Beer, M.; Schwemmle, M. A Genome-Wide CRISPR-Cas9 Screen Reveals the Requirement of Host Cell Sulfation for Schmallenberg Virus Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Labeau, A.; Simon-Loriere, E.; Hafirassou, M.-L.; Bonnet-Madin, L.; Tessier, S.; Zamborlini, A.; Dupré, T.; Seta, N.; Schwartz, O.; Chaix, M.-L.; et al. A Genome-Wide CRISPR-Cas9 Screen Identifies the Dolichol-Phosphate Mannose Synthase Complex as a Host Dependency Factor for Dengue Virus Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, K.; Bae, S.; Park, J.; Lee, C.K.; Kim, M.; Kim, E.; Kim, M.; Kim, S.; Kim, C.; et al. CRISPR/Cas9-mediated gene knockout screens and target identification via whole-genome sequencing uncover host genes required for picornavirus infection. J. Biol. Chem. 2017, 292, 10664–10671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulsuptrakul, J.; Wang, R.; Meyers, N.L.; Ott, M.; Puschnik, A.S. A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection. Cell Rep. 2021, 34, 108859. [Google Scholar] [CrossRef] [PubMed]
- Heaton, B.E.; Kennedy, E.M.; Dumm, R.E.; Harding, A.T.; Sacco, M.T.; Sachs, D.; Heaton, N.S. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep. 2017, 20, 1503–1512. [Google Scholar] [CrossRef] [Green Version]
- Moskovskich, A.; Goldmann, U.; Kartnig, F.; Lindinger, S.; Konecka, J.; Fiume, G.; Girardi, E.; Superti-Furga, G. The transporters SLC35A1 and SLC30A1 play opposite roles in cell survival upon VSV virus infection. Sci. Rep. 2019, 9, 10471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Liu, H.; Xiao, T.; Wang, Z.; Nie, X.; Li, X.; Qian, P.; Qin, L.; Han, X.; Zhang, J.; et al. CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication. Nat. Commun. 2020, 11, 5178. [Google Scholar] [CrossRef]
- Urbanek, K.; Sutherland, D.M.; Orchard, R.C.; Wilen, C.B.; Knowlton, J.J.; Aravamudhan, P.; Taylor, G.M.; Virgin, H.W.; Dermody, T.S. Cytidine Monophosphate N-Acetylneuraminic Acid Synthetase and Solute Carrier Family 35 Member A1 Are Required for Reovirus Binding and Infection. J. Virol. 2020, 95. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Ilan, N.; Naggi, A.; Casu, B. Heparanase: Structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr. Pharm. Des. 2007, 13, 2057–2073. [Google Scholar] [CrossRef] [PubMed]
- Hadigal, S.R.; Agelidis, A.M.; Karasneh, G.A.; Antoine, T.E.; Yakoub, A.M.; Ramani, V.C.; Djalilian, A.R.; Sanderson, R.D.; Shukla, D. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat. Commun. 2015, 6, 6985. [Google Scholar] [CrossRef] [Green Version]
- Lohse, D.L.; Linhardt, R.J. Purification and Characterization of Heparin Lyases from Flavobacterium-Heparinum. J. Biol. Chem. 1992, 267, 24347–24355. [Google Scholar] [CrossRef]
- Rivara, S.; Milazzo, F.M.; Giannini, G. Heparanase: A rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med. Chem. 2016, 8, 647–680. [Google Scholar] [CrossRef] [Green Version]
- Tseligka, E.D.; Sobo, K.; Stoppini, L.; Cagno, V.; Abdul, F.; Piuz, I.; Meylan, P.; Huang, S.; Constant, S.; Tapparel, C. A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PLoS Pathog. 2018, 14, e1007190. [Google Scholar] [CrossRef]
- Chu, H.; Hu, B.; Huang, X.; Chai, Y.; Zhou, D.; Wang, Y.; Shuai, H.; Yang, D.; Hou, Y.; Zhang, X.; et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Royston, L.; Essaidi-Laziosi, M.; Pérez-Rodríguez, F.J.; Piuz, I.; Geiser, J.; Krause, K.-H.; Huang, S.; Constant, S.; Kaiser, L.; Garcin, D.; et al. Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature. PLoS Pathog. 2018, 14. [Google Scholar] [CrossRef] [Green Version]
- Ciarlet, M.; Estes, M.K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 1999, 80, 943–948. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Aschenbrenner, L.M.; Smee, D.F.; Wandersee, M.K.; Sidwell, R.W.; Gubareva, L.V.; Mishin, V.P.; Hayden, F.G.; Kim, D.H.; Ing, A.; et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob. Agents Chemother. 2006, 50, 1470–1479. [Google Scholar] [CrossRef] [Green Version]
- Chibanga, V.P.; Dirr, L.; Guillon, P.; El-Deeb, I.M.; Bailly, B.; Thomson, R.J.; von Itzstein, M. New antiviral approaches for human parainfluenza: Inhibiting the haemagglutinin-neuraminidase. Antivir. Res. 2019, 167, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholls, J.M.; Moss, R.B.; Haslam, S.M. The use of sialidase therapy for respiratory viral infections. Antivir. Res. 2013, 98, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Esseili, M.A.; Gao, X.; Boley, P.; Hou, Y.; Saif, L.J.; Brewer-Jensen, P.; Lindesmith, L.C.; Baric, R.S.; Atmar, R.L.; Wang, Q. Human Norovirus Histo-Blood Group Antigen (HBGA) Binding Sites Mediate the Virus Specific Interactions with Lettuce Carbohydrates. Viruses 2019, 11, 833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Candelero-Rueda, R.A.; Saif, L.J.; Vlasova, A.N. Infection of porcine small intestinal enteroids with human and pig rotavirus A strains reveals contrasting roles for histo-blood group antigens and terminal sialic acids. PLoS Pathog. 2021, 17, e1009237. [Google Scholar] [CrossRef] [PubMed]
- Heida, R.; Bhide, Y.C.; Gasbarri, M.; Kocabiyik, O.; Stellacci, F.; Huckriede, A.L.W.; Hinrichs, W.L.J.; Frijlink, H.W. Advances in the development of entry inhibitors for sialic-acid-targeting viruses. Drug Discov. Today 2021, 26, 122–137. [Google Scholar] [CrossRef]
- Kocabiyik, O.; Cagno, V.; Silva, P.J.; Zhu, Y.; Sedano, L.; Bhide, Y.; Mettier, J.; Medaglia, C.; Da Costa, B.; Constant, S.; et al. Non-Toxic Virucidal Macromolecules Show High Efficacy against Influenza Virus Ex Vivo and In Vivo. Adv. Sci. 2021, 8, 2001012. [Google Scholar] [CrossRef]
- Ali, E.S.; Rajapaksha, H.; Carr, J.M.; Petrovsky, N. Norovirus drug candidates that inhibit viral capsid attachment to human histo-blood group antigens. Antivir. Res. 2016, 133, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.A.; Ramessar, K.; O’Keefe, B.R. Antiviral lectins: Selective inhibitors of viral entry. Antivir. Res 2017, 142, 37–54. [Google Scholar] [CrossRef]
- Hu, Y.; Meng, X.; Zhang, F.; Xiang, Y.; Wang, J. The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg. Microbes Infect. 2021, 10, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, C.; Wotring, J.W.; Zhang, C.J.; McCarty, S.M.; Fursmidt, R.; Frum, T.; Kadambi, N.S.; Amin, A.T.; O’Meara, T.R.; Pretto, C.D.; et al. Morphological Cell Profiling of SARS-CoV-2 Infection Identifies Drug Repurposing Candidates for COVID-19. BioRxiv 2020. [Google Scholar] [CrossRef]
- Cagno, V.; Andreozzi, P.; D’Alicarnasso, M.; Jacob Silva, P.; Mueller, M.; Galloux, M.; Le Goffic, R.; Jones, S.T.; Vallino, M.; Hodek, J.; et al. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 2018, 17, 195–203. [Google Scholar] [CrossRef]
- Jones, S.T.; Cagno, V.; Janecek, M.; Ortiz, D.; Gasilova, N.; Piret, J.; Gasbarri, M.; Constant, D.A.; Han, Y.; Vukovic, L.; et al. Modified cyclodextrins as broad-spectrum antivirals. Sci. Adv. 2020, 6, eaax9318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, C.F.; Tyssen, D.; Sonza, S.; Davie, A.; Evans, S.; Lewis, G.R.; Xia, S.; Spelman, T.; Hodsman, P.; Moench, T.R.; et al. SPL7013 Gel (VivaGel(R)) retains potent HIV-1 and HSV-2 inhibitory activity following vaginal administration in humans. PLoS ONE 2011, 6, e24095. [Google Scholar] [CrossRef] [Green Version]
- Magnan, S.; Tota, J.E.; El-Zein, M.; Burchell, A.N.; Schiller, J.T.; Ferenczy, A.; Tellier, P.P.; Coutlee, F.; Franco, E.L.; Group, C.S. Efficacy of a Carrageenan gel Against Transmission of Cervical HPV (CATCH): Interim analysis of a randomized, double-blind, placebo-controlled, phase 2B trial. Clin. Microbiol. Infect. 2019, 25, 210–216. [Google Scholar] [CrossRef] [Green Version]
- ClinicalTrial.gov. Lactoferrin Use in (SARS-CoV-2) Management. Available online: https://clinicaltrials.gov/ct2/results?pg=1&load=cart&id=NCT04860219 (accessed on 1 June 2021).
- Von Itzstein, M.; Wu, W.Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; Van Phan, T.; Smythe, M.L.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar] [CrossRef]
- Rusnati, M.; Chiodelli, P.; Bugatti, A.; Urbinati, C. Bridging the past and the future of virology: Surface plasmon resonance as a powerful tool to investigate virus/host interactions. Crit. Rev. Microbiol. 2015, 41, 238–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, A.; Valverde, P.; Arda, A.; Jimenez-Barbero, J. Glycan structures and their interactions with proteins. A NMR view. Curr. Opin. Struct. Biol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Koehler, M.; Delguste, M.; Sieben, C.; Gillet, L.; Alsteens, D. Initial Step of Virus Entry: Virion Binding to Cell-Surface Glycans. Ann. Rev. Virol. 2020, 7, 143–165. [Google Scholar] [CrossRef] [PubMed]
- Moulard, M.; Lortat-Jacob, H.; Mondor, I.; Roca, G.; Wyatt, R.; Sodroski, J.; Zhao, L.; Olson, W.; Kwong, P.D.; Sattentau, Q.J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J. Virol. 2000, 74, 1948–1960. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.K.; Straus, S.E. Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J. Virol. 1997, 71, 1375–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Jin, W.; Sood, A.; Montgomery, D.W.; Grant, O.C.; Fuster, M.M.; Fu, L.; Dordick, J.S.; Woods, R.J.; Zhang, F.; et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir. Res. 2020, 181, 104873. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Gandhi, N.S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar] [CrossRef] [PubMed]
- Schieble, J.H.; Fox, V.L.; Lennette, E.H. A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol. 1967, 85, 297–310. [Google Scholar] [CrossRef]
- Brown, D.M.; Hixon, A.M.; Oldfield, L.M.; Zhang, Y.; Novotny, M.; Wang, W.; Das, S.R.; Shabman, R.S.; Tyler, K.L.; Scheuermann, R.H. Contemporary Circulating Enterovirus D68 Strains Have Acquired the Capacity for Viral Entry and Replication in Human Neuronal Cells. MBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Hu, X.-Y.; Yu, X.-F. Current Understanding of Human Enterovirus D68. Viruses 2019, 11, 490. [Google Scholar] [CrossRef] [Green Version]
- Imamura, T.; Okamoto, M.; Nakakita, S.I.; Suzuki, A.; Saito, M.; Tamaki, R.; Lupisan, S.; Roy, C.N.; Hiramatsu, H.; Sugawara, K.E.; et al. Antigenic and Receptor Binding Properties of Enterovirus 68. J. Virol. 2014, 88, 2374–2384. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.-W.; Sun, M.; Guo, L.; Wang, J.-J.; Song, J.; Li, J.-Q.; Li, H.-Z.; Ning, R.-T.; Yang, Z.-N.; Fan, H.-T.; et al. Nasal Infection of Enterovirus D68 Leading to Lower Respiratory Tract Pathogenesis in Ferrets (Mustela putorius furo). Viruses 2017, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Hixon, A.M.; Clarke, P.; Tyler, K.L. Contemporary Circulating Enterovirus D68 Strains Infect and Undergo Retrograde Axonal Transport in Spinal Motor Neurons Independent of Sialic Acid. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sheng, J.; Baggen, J.; Meng, G.; Xiao, C.; Thibaut, H.J.; van Kuppeveld, F.J.M.; Rossmann, M.G. Sialic acid-dependent cell entry of human enterovirus D68. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, A.B.; Warren, A.L.; Racaniello, V.R. Neurotropism of Enterovirus D68 Isolates Is Independent of Sialic Acid and Is Not a Recently Acquired Phenotype. MBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Guo, H.; Chang, J.; Yu, Y.; Liu, G.; Zhang, N.; Willard, S.H.; Zheng, S.; Yu, X.-F. ICAM-5/Telencephalin Is a Functional Entry Receptor for Enterovirus D68. Cell Host Microbe 2016, 20, 631–641. [Google Scholar] [CrossRef] [Green Version]
- Cagno, V. SARS-CoV-2 cellular tropism. Lancet Microbe 2020, 1, e2–e3. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Severe Covid, G.G.; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Hao, W.; Ma, B.; Li, Z.; Wang, X.; Gao, X.; Li, Y.; Qin, B.; Shang, S.; Cui, S.; Tan, Z. Binding of the SARS-CoV-2 Spike Protein to Glycans. Sci. Bull. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020, 6, 80. [Google Scholar] [CrossRef]
- Gasbarri, M.; V’Kovski, P.; Torriani, G.; Thiel, V.; Stellacci, F.; Tapparel, C.; Cagno, V. SARS-CoV-2 Inhibition by Sulfonated Compounds. Microorganisms 2020, 8, 1894. [Google Scholar] [CrossRef]
- Kwon, P.S.; Oh, H.; Kwon, S.J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 50. [Google Scholar] [CrossRef]
- Ryzhikov, A.B.; Onkhonova, G.S.; Imatdinov, I.R.; Gavrilova, E.V.; Maksyutov, R.A.; Gordeeva, E.A.; Pazynina, G.V.; Ryzhov, I.M.; Shilova, N.V.; Bovin, N.V. Recombinant SARS-CoV-2 S Protein Binds to Glycans of the Lactosamine Family in vitro. Biochemistry 2021, 86, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef]
- Zhang, L.; Bukreyev, A.; Thompson, C.I.; Watson, B.; Peeples, M.E.; Collins, P.L.; Pickles, R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005, 79, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, S.M.; McNally, B.A.; Ioannidis, I.; Flano, E.; Teng, M.N.; Oomens, A.G.; Walsh, E.E.; Peeples, M.E. Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS Pathog. 2015, 11, e1005318. [Google Scholar] [CrossRef] [Green Version]

| Virus | Pathway Identified | Factor Identified | Reference |
|---|---|---|---|
| Chikungunya Virus | heparan sulfate | EXT1, EXT2, EXTL3, FAM20B, B3GAT3 | [32] |
| Encephalomyocarditis Virus | sialic acid | SLC35A1, CMAS | [33] |
| heparan sulfate | B3GAT3, SLC35B2 | ||
| Enterovirus D68 | sialic acid | GNE, NANS, CMAS, SLC35A1, SLC35A2, MGAT5, B4GALT1, ST3GAL4, ST6GAL1 | [29] |
| heparan sulfate | B3GAT3, FAM20B, B3GALT6, B4GALT7, UXS1, XYLT2, EXT1, EXT2, EXTL3, UGP2, UGDH, SLC35B2, NDST1 | [34] | |
| Lassa Virus | sialic acid | SLC35A1, CMAS, SLC35A2, GNE | [26] |
| N-glycosylation | ALG8, MAN1B1, ALG6, ALG5, MAN1A1, MGAT1 | ||
| α-dystroglycan glycosylation | LARGE, ISPD, FKTN, FKRP, POMT1, POMT2, DPM3, C3orf39all | ||
| Rift Valley Fever Virus | heparan sulfate | XYLT2, B4GALT7, B3GALT6, B3GAT3, EXTL3, EXT1, EXT2, NDST1, UXS1, UGDH, SLC35B2, PTAR1 | [28] |
| Vaccinia Virus | heparan sulfate | XYLT2, B4GALT7, B3GALT6, B3GAT3, EXTL3, EXT1, EXT2, HS2ST1, NDST1, UGDH, UXS1, SLC35B2, PTAR1 | [27] |
| Virus | Pathway Identified | Factor Identified | Reference |
|---|---|---|---|
| Dengue Virus | heparan sulfate | EXTL3, EXT2, B4GALT7, B3GALT6, B3GAT3, PAPSS1, SLC35B2 | [40] |
| dolichol-phosphate mannose synthetase | DPM1, DPM3 | ||
| Enterovirus D68 | sialic acid | ST3GAL4 | [41] |
| Hepatitis A virus | sialic acid | SLC35A1, UGCG, ST3GAL5, GNE, CMAS | [42] |
| Influenza H1N1 | sialic acid | B4GALNT2 | [43] |
| SLC35A1, SLC35A2 | [44] | ||
| Influenza H5N1 | sialic acid | GNE, CMAS, SLC35A1, SLC35A2, GANAB, ALG12, ALG3, DPM2, ALG5 | [36] |
| glycan modification | A4GALT, B3GAT1, B4GALNT4, CHSY1, PIGN, CSGALNACT2, HS3ST6 | ||
| Japanese encephalitis virus | heparan sulfate | EXT1, EXT2, GLCE, HS6ST1, B3GAT3, B4GALT7, XYLT7, EXTL3, SLC35B2, GAA | [45] |
| Reovirus | sialic acid | NANS, ST3GAL4, SLC35A1, CMAS | [46] |
| Schmallenberg Virus | heparan sulfate | SLC35B2 | [39] |
| Sindbis Virus | heparan sulfate | SLC35B2, B4GALT7, EXT2, EXT1 | [38] |
| Vesicular Stomatitis Virus | sialic acid | SLC35A1 | [44] |
| Zika Virus | heparan sulfate | TM9SF2, EXTL3, EXT2, NDST1, SLC35B2, EXT1, B4GALT7, PAPSS1, B3GALT6, HS6ST1 | [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathez, G.; Cagno, V. Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment. Microorganisms 2021, 9, 1238. https://doi.org/10.3390/microorganisms9061238
Mathez G, Cagno V. Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment. Microorganisms. 2021; 9(6):1238. https://doi.org/10.3390/microorganisms9061238
Chicago/Turabian StyleMathez, Gregory, and Valeria Cagno. 2021. "Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment" Microorganisms 9, no. 6: 1238. https://doi.org/10.3390/microorganisms9061238
APA StyleMathez, G., & Cagno, V. (2021). Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment. Microorganisms, 9(6), 1238. https://doi.org/10.3390/microorganisms9061238