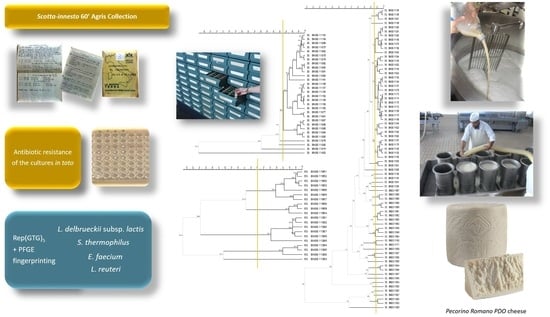
Biodiversity and Safety Assessment of Half-Century Preserved Natural Starter Cultures for Pecorino Romano PDO Cheese
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Reactivation of the Cultures
2.3. Microbial Counts and Isolation
2.4. Molecular Identification and Biotyping of the SR Isolates
2.5. Evaluation of Phenotypic Antibiotic Resistance of the Starter Cultures
2.6. Molecular Analysis for the Safety Evaluation and Identification of SR56 Isolates
3. Results and Discussion
3.1. Microbial Counts, Molecular Identification and Biotyping of the SR Isolates
3.2. Antibiotic Resistance in the Starter Cultures and Safety Assessment of Enterococcus Faecium Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Addis, M.; Fiori, M.; Riu, G.; Pes, M.; Salvatore, E.; Pirisi, A. Physico-chemical characteristics and acidic profile of PDO Pecorino Romano cheese: Seasonal variation. Small Rumin. Res. 2015, 126, 73–79. [Google Scholar] [CrossRef]
- Romano, C.p.l.t.d.f.P. Il Monitoraggio delle Vendite Presso la Distribuzione Moderna: Pecorino Romano DOP. Available online: https://www.pecorinoromano.com/application/files/8415/7323/4642/Pecorino_romano_DOP_Gennaio_Ago_2019_Ismea.pdf (accessed on 30 May 2021).
- CLAL.it. Italia: Export Pecorino e Fiore Sardo. Available online: https://www.clal.it/index.php?section=imp_exp_istat&cod=04069063&mov=E (accessed on 30 May 2021).
- CLAL.it. Italia: Produzioni di Pecorino Romano DOP. Available online: https://www.clal.it/index.php?section=produzioni_pecorino (accessed on 26 April 2021).
- EC No. 852/2004. In On the Hygiene of Foodstuffs; Official Journal of the European Union: Strasbourg, France, 2004.
- EC No. 510/2006. In On the Protection of Geographical Indications and Designations of Origin for Agricultural Products and Foodstuffs; Official Journal of the European Union: Brussels, Belgium, 2006.
- EC No. 1030/2009. In Approving Minor Amendments to the Specification of a Name Registered in the Register of Protected Designations of Origin and Protected Geographical Indications (Pecorino Romano (PDO); Official Journal of the European Union: Brussels, Belgium, 2009.
- IFCQ Certificazioni. Piano di Controllo Pecorino Romano DOP. PC PR Rev. 7 del 13 Dicembre 2011. Available online: https://ifcq.it/files/6d9eef87812362678d01a1935289b96cc9d46f10.pdf?action=download (accessed on 21 June 2021).
- De Vero, L.; Boniotti, M.B.; Budroni, M.; Buzzini, P.; Cassanelli, S.; Comunian, R.; Gullo, M.; Logrieco, A.F.; Mannazzu, I.; Musumeci, R.; et al. Preservation, Characterization and Exploitation of Microbial Biodiversity: The Perspective of the Italian Network of Culture Collections. Microorganisms 2019, 7, 685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Tech. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Chessa, L.; Paba, A.; Daga, E.; Caredda, M.; Comunian, R. Optimization of scotta as growth medium to preserve biodiversity and maximise bacterial cells concentration of natural starter cultures for Pecorino Romano PDO cheese. FEMS Microbiol. Lett. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chessa, L.; Paba, A.; Daga, E.; Comunian, R. Effect of growth media on natural starter culture composition and performance evaluated with a polyphasic approach. Int. J. Dairy Technol. 2019, 72, 152–158. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Additives Products or Substances Used in Animal Feed; Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.D.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef] [PubMed]
- Isolini, D.; Grand, M.; Glättli, H. Selektivmedien zum Nachweis von obligat und fakultativ heterofermen- tativen Laklobazillen. Schweiz Milchw. Forsch. 1990, 19, 57–59. [Google Scholar]
- Tosi, L.; Berruti, G.; Danielsen, M.; Wind, A.; Huys, G.; Morelli, L. Susceptibility of Streptococcus thermophilus to antibiotics. Antonie Leeuwenhoek 2007, 92, 21–28. [Google Scholar] [CrossRef]
- Graves, L.M.; Swaminathan, B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- Gosiewski, T.; Brzychczy-Wloch, M. The use of PFGE method in genotyping of selected bacteria species of the Lactobacillus genus. Methods Mol. Biol. 2015, 1301, 225–240. [Google Scholar] [CrossRef]
- ISO 10932:2010 [IDF 223:2010]. In Milk and Milk Products in Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB); ISO 10932:2010; International Organization for Standardization (ISO): Geneva, Switzerland, 2010.
- Clinical and Laboratory Standards Institute (CLSI). M100—Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; United State: Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- The National Antimicrobial Resistance Monitoring System: NARMS Integrated Report 2016–2017; Department of Health and Human Services, FDA: Laurel, MD, USA, 2019.
- Comunian, R.; Paba, A.; Daga, E.; Dupré, I.; Scintu, M.F. Traditional and innovative production methods of Fiore Sardo cheese: A comparison of microflora with a PCR-culture technique. Int. J. Dairy Technol. 2010, 63, 224–233. [Google Scholar] [CrossRef]
- EFSA Panel on Additives Products or Substances used in Animal Feed. Guidance on the safety assessment of Enterococcus faecium in animal nutrition. EFSA J. 2012, 10, 2682. [Google Scholar] [CrossRef]
- Lazzi, C.; Rossetti, L.; Zago, M.; Neviani, E.; Giraffa, G. Evaluation of bacterial communities belonging to natural whey starters for Grana Padano cheese by length heterogeneity-PCR. J. Appl. Microbiol. 2004, 96, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Parente, E.; Cogan, T. Starter Cultures: General Aspects. Cheese Chem. Phys. Microbiol. 2004, 1, 123–147. [Google Scholar]
- Galistu, G.; Piredda, G.; Pirisi, A.; Scintu, M.F.; Ledda, A. Pecorino Romano: Hard cooked ewe’s milk cheese. In Proceedings of the International Symposium on Basis of the Quality of Typical Mediterranean Animal Products, Zafra, Badajoz, Spain, 29 September–2 October 1996. [Google Scholar]
- Gobbetti, M.; Di Cagno, R. CHEESE|Hard Italian Cheeses. In Encyclopedia of Dairy Sciences; Roginski, H., Ed.; Elsevier: Oxford, UK, 2002. [Google Scholar] [CrossRef]
- Bottazzi, V.; Ledda, A. Microbiologia del formaggio pecorino “romano”. Ann. Micr. 1967, 17, 41–53. [Google Scholar]
- Foulquié Moreno, M.R.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Pirisi, A.; Comunian, R.; Urgeghe, P.P.; Scintu, M.F. Sheep’s and goat’s dairy products in Italy: Technological, chemical, microbiological, and sensory aspects. Small Rumin. Res. 2011, 101, 102–112. [Google Scholar] [CrossRef]
- Schirru, S.; Todorov, S.D.; Favaro, L.; Mangia, N.P.; Basaglia, M.; Casella, S.; Comunian, R.; Franco, B.D.G.D.M.; Deiana, P. Sardinian goat’s milk as source of bacteriocinogenic potential protective cultures. Food Control. 2012, 25, 309–320. [Google Scholar] [CrossRef]
- Dal Bello, F.; Walter, J.; Roos, S.; Jonsson, H.; Hertel, C. Inducible Gene Expression in Lactobacillus reuteri LTH5531 during Type II Sourdough Fermentation. Appl. Environ. Microbiol. 2005, 71, 5873–5878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casas, I.A.; Dobrogosz, W.J. Validation of the Probiotic Concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 2000, 12, 247–285. [Google Scholar] [CrossRef] [Green Version]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Shornikova, A.V.; Casas, I.A.; Mykkänen, H.; Salo, E.; Vesikari, T. Bacteriotherapy with Lactobacillus reuteri in rotavirus gastroenteritis. Pediatric Infect. Dis. J. 1997, 16, 1103–1107. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, C.; Huang, J.; Kuai, X.; Shao, X. The potential therapeutic role of Lactobacillus reuteri for treatment of inflammatory bowel disease. Am. J. Transl Res. 2020, 12, 1569–1583. [Google Scholar] [PubMed]
- el-Ziney, M.G.; Debevere, J.M. The effect of Reuterin on Listeria monocytogenes and Escherichia coli O157:H7 in milk and cottage cheese. J. Food Prot. 1998, 61, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Arqués, J.L.; Fernández, J.; Gaya, P.; Nuñez, M.; Rodríguez, E.; Medina, M. Antimicrobial activity of reuterin in combination with nisin against food-borne pathogens. Int. J. Food Microbiol. 2004, 95, 225–229. [Google Scholar] [CrossRef]
- Morandi, S.; Brasca, M.; Andrighetto, C.; Lombardi, A.; Lodi, R. Technological and molecular characterisation of enterococci isolated from north–west Italian dairy products. Int. Dairy J. 2006, 16, 867–875. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti. Infect. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- Mrkonjic Fuka, M.; Zgomba Maksimovic, A.; Tanuwidjaja, I.; Hulak, N.; Schloter, M. Characterization of Enterococcal Community Isolated from an Artisan Istrian Raw Milk Cheese: Biotechnological and Safety Aspects. Food Technol. Biotechnol. 2017, 55, 368–380. [Google Scholar] [CrossRef]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Dapkevicius, M.D.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current Trends of Enterococci in Dairy Products: A Comprehensive Review of Their Multiple Roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, Ma, USA, 2014. [Google Scholar]
- Haghi, F.; Lohrasbi, V.; Zeighami, H. High incidence of virulence determinants, aminoglycoside and vancomycin resistance in enterococci isolated from hospitalized patients in Northwest Iran. BMC Infect. Dis. 2019, 19, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kak, V.; Chow, J.W. Acquired Antibiotic Resistances in Enterococci. In The Enterococci; American Society for Microbiology: Washington, DC, USA, 2014; pp. 355–383. [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical Microbiology Infectious Diseases. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [Google Scholar] [CrossRef] [Green Version]
- Leuschner, R.G.K.; Robinson, T.P.; Hugas, M.; Cocconcelli, P.S.; Richard-Forget, F.; Klein, G.; Licht, T.R.; Nguyen-The, C.; Querol, A.; Richardson, M.; et al. Qualified presumption of safety (QPS): A generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA). Trends Food Sci. Technol. 2010, 21, 425–435. [Google Scholar] [CrossRef]
- van Reenen, C.A.; Dicks, L.M. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: What are the possibilities? A review. Arch. Microbiol. 2011, 193, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Fleige, C.; Geringer, U.; van Schaik, W.; Klare, I.; Witte, W. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 2011, 11, 80. [Google Scholar] [CrossRef] [Green Version]
- Rice, L.B.; Carias, L.; Rudin, S.; Vael, C.; Goossens, H.; Konstabel, C.; Klare, I.; Nallapareddy, S.R.; Huang, W.; Murray, B.E. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 2003, 187, 508–512. [Google Scholar] [CrossRef] [Green Version]
- Aun, E.; Kisand, V.; Laht, M.; Telling, K.; Kalmus, P.; Väli, Ü.; Brauer, A.; Remm, M.; Tenson, T. Molecular Characterization of Enterococcus Isolates From Different Sources in Estonia Reveals Potential Transmission of Resistance Genes Among Different Reservoirs. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Cheng, G.; Ning, J.; Ahmed, S.; Huang, J.; Ullah, R.; An, B.; Hao, H.; Dai, M.; Huang, L.; Wang, X.; et al. Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control. 2019, 8, 158. [Google Scholar] [CrossRef] [Green Version]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [Green Version]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.-M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schjørring, S.; Krogfelt, K.A. Assessment of Bacterial Antibiotic Resistance Transfer in the Gut. Int. J. Microbiol. 2011, 2011, 312956. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Antibiotic | Concentration | Culture Medium | Microbial Culture | ||
|---|---|---|---|---|---|
| (mg/L) | SR30 | SR56 | SR63 | ||
| Amp | 2 1 | LSM | − | − | − |
| ISTL | n.t. | n.t. | n.t. | ||
| 4 | LSM | − | − | − | |
| ISTL | − | − | − | ||
| 8 2 | LSM | − | − | − | |
| ISTL | n.t. | n.t. | n.t. | ||
| Chl | 8 1,2 | LSM | − | − | − |
| ISTL | − | − | − | ||
| 16 | LSM | − | − | − | |
| ISTL | n.t. | n.t. | n.t. | ||
| 32 | LSM | n.t. | n.t. | n.t. | |
| ISTL | − | − | − | ||
| Cli 2 | 2 | LSM | + | + | − |
| ISTL | n.t. | n.t. | n.t. | ||
| 4 1 | LSM | n.t. | n.t. | n.t. | |
| ISTL | + | + | − | ||
| 8 | LSM | n.t. | n.t. | n.t. | |
| ISTL | − | + | − | ||
| Ery | 1 | LSM | + | + | − |
| ISTL | + | + | − | ||
| 2 | LSM | − | + | − | |
| ISTL | − | + | − | ||
| 4 1,2 | LSM | − | + | − | |
| ISTL | − | + | − | ||
| 8 | LSM | − | + | − | |
| ISTL | − | + | − | ||
| Gen | 16 | LSM | − | + | + |
| ISTL | n.t. | n.t. | n.t. | ||
| 32 1 | LSM | − | + | − | |
| ISTL | − | − | − | ||
| Tet | 4 1 | LSM | − | + | − |
| ISTL | − | + | − | ||
| 8 2 | LSM | − | + | − | |
| ISTL | − | + | − | ||
| 16 | LSM | − | + | − | |
| ISTL | − | − | − | ||
| Til | 8 3 | LSM | − | − | − |
| ISTL | − | − | − | ||
| Antibiotic | Concentration | Culture Medium | Microbial Dilution 1 | ||
|---|---|---|---|---|---|
| (mg/L) | 1/10 | 1/100 | 1/1000 | ||
| Ery | 2 | LSM | + | + | + |
| ISTL | + | − | − | ||
| 4 | LSM | + | + | + | |
| ISTL | + | − | − | ||
| 8 | LSM | + | + | − | |
| ISTL | + | − | − | ||
| Tet | 4 | LSM | + | + | + |
| ISTL | + | + | + | ||
| 8 | LSM | + | + | + | |
| ISTL | + | + | + | ||
| 16 | LSM | − | − | − | |
| ISTL | − | − | − | ||
| Biodiversity | Safety Assessment 1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cultures | Species | Log CFU/mL | Number of Isolates | Number of Biotypes | Phenotypic Antibiotic Susceptibility 2 | Molecular Antibiotic Resistance 3,4 | E. faecium Isolates 5 Specific Tests [13] | |||||
| Cultures in toto | CFU/mL Estimation | Cultures in toto | Colonies | Amp Resistance | esp | hylefm | IS16 | |||||
| SR30 | Amp Neg Chl Neg Cli 4 mg/L Ery 1 mg/L Gen Neg Tet Neg Til Neg | n.t. | n.t. | n.t. | <2 mg/L | Neg | Neg | Neg | ||||
| S. thermophilus | 8.72 | 30 | 5 | |||||||||
| E. faecium | 6.32 | 45 | 16 | |||||||||
| SR56 | Amp Neg Chl Neg Cli 8 mg/L Ery 8 mg/L Gen 32 mg/L Tet 16 mg/L Til Neg | Ery2/4 mg/L: 4 Log Ery8 mg/L: 3 Log Tet4/8 mg/L: 4 Log | tetM | tetM | <2 mg/L | Neg | Neg | Neg | ||||
| E. faecium | 8.03 | 24 | 20 | |||||||||
| L. delbrueckii lactis | 9.41 | 40 | 17 | |||||||||
| SR63 | Amp Neg Chl Neg Cli Neg Ery Neg Gen 16 mg/L Tet Neg Til Neg | n.t. | n.t. | n.t. | <2 mg/L | Neg | Neg | Neg | ||||
| E. faecium | 2.56 | 18 | 7 | |||||||||
| L. delbrueckii lactis | 8.67 | 32 | 18 | |||||||||
| L. reuteri | 4.18 | 20 | 8 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chessa, L.; Paba, A.; Daga, E.; Dupré, I.; Comunian, R. Biodiversity and Safety Assessment of Half-Century Preserved Natural Starter Cultures for Pecorino Romano PDO Cheese. Microorganisms 2021, 9, 1363. https://doi.org/10.3390/microorganisms9071363
Chessa L, Paba A, Daga E, Dupré I, Comunian R. Biodiversity and Safety Assessment of Half-Century Preserved Natural Starter Cultures for Pecorino Romano PDO Cheese. Microorganisms. 2021; 9(7):1363. https://doi.org/10.3390/microorganisms9071363
Chicago/Turabian StyleChessa, Luigi, Antonio Paba, Elisabetta Daga, Ilaria Dupré, and Roberta Comunian. 2021. "Biodiversity and Safety Assessment of Half-Century Preserved Natural Starter Cultures for Pecorino Romano PDO Cheese" Microorganisms 9, no. 7: 1363. https://doi.org/10.3390/microorganisms9071363
APA StyleChessa, L., Paba, A., Daga, E., Dupré, I., & Comunian, R. (2021). Biodiversity and Safety Assessment of Half-Century Preserved Natural Starter Cultures for Pecorino Romano PDO Cheese. Microorganisms, 9(7), 1363. https://doi.org/10.3390/microorganisms9071363