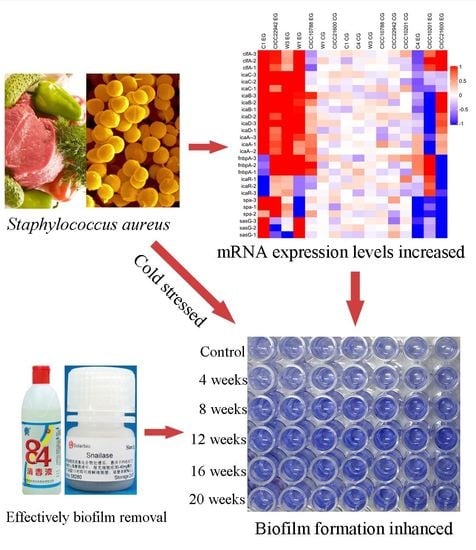
Research on the Biofilm Formation of Staphylococcus aureus after Cold Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Biofilm Formation of S. aureus Cells with Cold Stress
2.3. The Growth of S. aureus Biofilm Cells after Cold Stress
2.4. Relative Transcription Levels of Biofilm-Associated Genes
2.4.1. RNA Extraction
2.4.2. Reverse Transcription
2.4.3. RT-PCR
2.5. Detection of Extracellular Polysaccharides in Biofilm Matrix
2.6. Evaluation of Protein in Biofilm Matrix
2.7. Hydrophobicity of S. aureus Strains after Cold Stress
2.8. Antibiotics and Enzymes to Remove S. aureus Biofilm
2.9. The Proteinase K, Snailase, or 84 Disinfectant Degraded S. aureus Biofilm
2.10. Statistical Analysis
3. Results
3.1. Biofilm Formation of Staphylococcus aureus Strains after Cold Stress
3.2. The Growth of S. aureus Biofilm Cells after Cold Stress
3.3. Transcription of Genes in Staphylococcus aureus Related to Biofilm Formation
3.4. The Polysaccharide and Protein Content in the Biofilm Matrix of S. aureus Cells
3.5. Hydrophobicity of S. aureus Strains after Cold Stress
3.6. Antibiotics and Enzymes Degraded S. aureus Biofilm
3.7. The Proteinase K, Snailase, or 84 Disinfectant Degraded S. aureus Biofilm
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jamali, H.; Paydar, M.; Radmehr, B.; Ismail, S.; Dadrasnia, A. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control. 2015, 54, 383–388. [Google Scholar] [CrossRef]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 2016, 22, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [Green Version]
- Fetsch, A.; Contzen, M.; Hartelt, K.; Kleiser, A.; Maassen, S.; Rau, J.; Kraushaar, B.; Layer, F.; Strommenger, B. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int. J. Food Microbiol. 2014, 187, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Puah, S.M.; Chua, K.H.; Tan, J.A. Virulence factors and antibiotic susceptibility of Staphylococcus aureus isolates in ready-to-eat foods: Detection of S. aureus contamination and a high prevalence of virulence genes. Int. J. Environ. Res. Public Health 2016, 13, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.Q.; Gao, X.P.; Zhang, Q.H.; Liu, Y.X.; Zhao, G.M.; Miao-Yun, L.I.; Sun, L.X.; Zhang, J.W. Total bacte-rial counts in fast-frozen dumplings and rice balls produced in different months. Food Sci. 2011, 32, 293–295. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Wang, X.; Tao, X.; Xia, X.; Yang, B.; Xi, M.; Meng, J.; Zhang, J.; Xu, B. Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail raw chicken in China. Food Control. 2013, 29, 103–106. [Google Scholar] [CrossRef]
- Hou, W.; Sun, X.; Wang, Z.; Zhang, Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5624–5631. [Google Scholar] [CrossRef] [Green Version]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [Green Version]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Hisatsune, J.; Hayashi, I.; Tatsukawa, N.; Sato’o, Y.; Mizumachi, E.; Hirakawa, H.; Pier, G.B.; Sugai, M.; Kato, F. A Novel repressor of the ica locus discovered in clinically isolated super-biofilm-elaborating Staphylococcus aureus. mBio 2017, 8, e02282-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arciola, C.R.; Campoccia, D.; Speziale, P.; Montanaro, L.; Costerton, J.W. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 2012, 33, 5967–5982. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, A.; Langsrud, S.; Heir, E.; Mikkelsen, M.I.; Moretro, T. Biofilm matrix composition affects the susceptibility of food associated Staphylococci to cleaning and disinfection agents. Front. Microbiol. 2016, 7, 856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rode, T.M.; Langsrud, S.; Holck, A.; Moretro, T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int. J. Food Microbiol. 2007, 116, 372–383. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C. Stress adaptation in foodborne pathogens. Annu. Rev. Food Sci. Technol. 2015, 6, 191–210. [Google Scholar] [CrossRef]
- Ricke, S.C.; Dawoud, T.M.; Kim, S.A.; Park, S.H.; Kwon, Y.M. Salmonella Cold Stress Response: Mechanisms and Occurrence in Foods. Int. Rev. Cytol. 2018, 104, 1–38. [Google Scholar]
- Lee, B.H.; Hebraud, M.; Bernardi, T. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol. 2017, 8, 2221. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Pratt, L.A.; Watnick, P.I.; Newman, D.K.; Weaver, V.B.; Kolter, R. Genetic approaches to study of biofilms. Methods Enzymol. 1999, 310, 91–109. [Google Scholar]
- Asadishad, B.; Olsson, A.L.; Dusane, D.H.; Ghoshal, S.; Tufenkji, N. Transport, motility, biofilm forming potential and survival of Bacillus subtilis exposed to cold temperature and freeze-thaw. Water Res. 2014, 58, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Qiao, J.; Zhu, M.; Lu, Z.; Lv, F.; Bie, X. The antibiotics resistance mechanism and pathogenicity of cold stressed Staphylococcus aureus. LWT 2020, 126, 109274. [Google Scholar] [CrossRef]
- Qiao, J.; Zhu, M.; Fan, Y.; Lu, Z.; Lv, F.; Zhao, H.; Bie, X. Properties and control of cold-induced small colony variants of Staphylococcus aureus. Food Biosci. 2021, 40, 100874. [Google Scholar] [CrossRef]
- Tahmasebi, H.; Dehbashi, S.; Arabestani, M.R. New approach to identify colistin-resistant Pseudomonas aeruginosa by high-resolution melting curve analysis assay. Lett. Appl. Microbiol. 2020, 70, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Shiesh, S.-C.; Wu, S.-J. Rapid detection of K-ras mutations in bile by peptide nucleic acid-mediated PCR clamping and melting curve analysis: Comparison with restriction fragment length polymorphism analysis. Clin. Chem. 2004, 50, 481–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genom. 2014, 15, 648. [Google Scholar] [CrossRef] [Green Version]
- Peirson, S.N.; Butler, J.N.; Foster, R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003, 31, e73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Wu, X.; Li, J.; Liu, L.; Zhang, R.; Shao, D.; Du, X. The specific anti-biofilm effect of gallic acid on Staphylococcus aureus by regulating the expression of the ica operon. Food Control. 2017, 73, 613–618. [Google Scholar] [CrossRef]
- Li, P.; Chen, X.; Shen, Y.; Li, H.; Zou, Y.; Yuan, G.; Hu, P.; Hu, H. Mucus penetration enhanced lipid polymer nanoparticles improve the eradication rate of Helicobacter pylori biofilm. J. Control. Release 2019, 300, 52–63. [Google Scholar] [CrossRef]
- Angiolella, L.; Leone, C.; Rojas, F.; Mussin, J.; de Los Angeles Sosa, M.; Giusiano, G. Biofilm, adherence, and hydrophobicity as virulence factors in Malassezia furfur. Med. Mycol. 2018, 56, 110–116. [Google Scholar] [CrossRef]
- Ruhs, P.A.; Bocker, L.; Inglis, R.F.; Fischer, P. Studying bacterial hydrophobicity and biofilm formation at liquid-liquid interfaces through interfacial rheology and pendant drop tensiometry. Colloids Surf. B Biointerfaces 2014, 117, 174–184. [Google Scholar] [CrossRef]
- Miao, J.; Liang, Y.; Chen, L.; Wang, W.; Wang, J.; Li, B.; Li, L.; Chen, D.; Xu, Z. Formation and development of Staphylococcus biofilm: With focus on food safety. J. Food Saf. 2017, 37, e12358. [Google Scholar] [CrossRef]
- De Souza, E.L.; Meira, Q.G.S.; Barbosa, I.D.M.; Athayde, A.J.A.A.; Da Conceição, M.L.; Júnior, J.P.D.S. Biofilm formation by Staphylococcus aureus from food contact surfaces in a meat-based broth and sensitivity to sanitizers. Braz. J. Microbiol. 2014, 45, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llorens, A.; Lloret, E.; Picouet, P.A.; Trbojevich, R.; Fernandez, A. Metallic-based micro and nanocomposites in food contact materials and active food packaging. Trends Food Sci. Technol. 2012, 24, 19–29. [Google Scholar] [CrossRef]
- Miao, J.; Lin, S.; Soteyome, T.; Peters, B.M.; Li, Y.; Chen, H.; Su, J.; Li, L.; Li, B.; Xu, Z.; et al. Biofilm formation of Staphylococcus aureus under food heat processing conditions: First report on CML produc-tion within biofilm. Sci. Rep. 2019, 9, 1312. [Google Scholar] [CrossRef]
- Møretrø, T.; Langsrud, S.; Heir, E. Bacteria on meat abattoir process surfaces after sanitation: Characterisation of survival properties of Listeria monocytogenes and the commensal bacterial flora. Adv. Microbiol. 2013, 3, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Rossi, E.; Paroni, M.; Landini, P. Biofilm and motility in response to environmental and host-related signals in Gram negative opportunistic pathogens. J. Appl. Microbiol. 2018, 125, 1587–1602. [Google Scholar] [CrossRef] [Green Version]
- Meira, Q.G.D.S.; Barbosa, I.D.M.; Athayde, A.J.A.A.; Siqueira-Júnior, J.P.D.; Souza, E.L.D. Influence of tem-perature and surface kind on biofilm formation by Staphylococcus aureus from food-contact surfaces and sensitivity to sanitizers. Food Control. 2012, 25, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Gerke, C.; Kraft, A.; Sussmuth, R.; Schweitzer, O.; Gotz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesion *. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2010, 6, 269–275. [Google Scholar] [CrossRef]
- Rohde, H.; Frankenberger, S.; Zahringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.A.; Corrigan, R.M.; Gruszka, D.; Speziale, P.; O’Gara, J.P.; Potts, J.R.; Foster, T.J. Role of Surface Protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2010, 192, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penadeés, J.R.; Lasa, I. Protein A-Mediated Multicellular Behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, T.; Foster, T.J.; O’Gara, J.P. A Novel Staphylococcus aureus Biofilm Phenotype Mediated by the Fibronectin-Binding Proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizan, M.F.R.; Jahid, I.K.; Kim, M.; Lee, K.-H.; Kim, T.J.; Ha, S.-D. Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling 2016, 32, 497–509. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- K alya, A.V.; Ahearn, D.G. Increased resistance to antifungal antibiotics of Candida spp. adhered to silicone. J. Ind. Microbiol. Biotechnol. 1995, 14, 451–455. [Google Scholar]
- Samaranayake, Y.; Cheung, B.; Yau, J.; Yeung, S.; Samaranayake, L. Human Serum Promotes Candida albicans Biofilm Growth and Virulence Gene Expression on Silicone Biomaterial. PLoS ONE 2013, 8, e62902. [Google Scholar] [CrossRef] [Green Version]
- R an, S.J.; Jiang, W.; Zhu, C.L.; Liang, J.P. Exploration of the mechanisms of biofilm formation by Enterococcus faecalis in glucose starvation environments. Aust. Dent. J. 2015, 60, 143–153. [Google Scholar] [CrossRef]
- Tyfa, A.; Kunicka-Styczynska, A.; Zabielska, J. Evaluation of hydrophobicity and quantitative analysis of biofilm formation by Alicyclobacillus sp. Acta Biochim. Pol. 2015, 62, 785–790. [Google Scholar] [CrossRef]
- Gilbert, P.; Allison, D.G.; McBain, A. Biofilms in vitro and in vivo: Do singular mechanisms imply cross-resistance? J. Appl. Microbiol. Symp. Suppl. 2002, 92, 98–110. [Google Scholar] [CrossRef]
- Romaní, A.M.; Fund, K.; Artigas, J.; Schwartz, T.; Sabater, S.; Obst, U. Relevance of polymeric matrix enzymes during biofilm formation. Microb. Ecol. 2008, 56, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Yan g, Y.L.; Chen, M.; Gu, J.L.; Zhu, F.Y.; Xu, X.G.; Zhang, C.; Chen, J.H.; Pan, W.H.; Liao, W.Q. Cryptococcosis in kidney transplant recipients in a Chinese university hospital and a review of published cases. Int. J. Infect. Dis. 2014, 26, 154–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suri, D.R. The use of human Deoxyribonuclease (rhDNase) in the management of cystic fibrosis. Biodrugs. 2005, 19, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and Antibiotics on Biofilm Characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irit, G.; Alex, S. Effect of proteases on biofilm formation of the plastic-degrading actinomycete Rhodococcus ruber C208. FEMS Microbiol. Lett. 2013, 342, 18–23. [Google Scholar]
- Elchinger, P.H.; Delattre, C.; Faure, S.; Roy, O.; Badel, S.; Bernardi, T.; Taillefumier, C.; Michaud, P. Effect of pro-teases against biofilms of Staphylococcus aureus and Staphylococcus epidermidis. Lett. Appl. Microbiol. 2015, 59, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Liu, J.; Gao, W. Effects of Snailase Treatment on Wettability of Raw Cotton Yarns in Pre-wetting Process of Foam Sizing. Appl. Biochem. Biotechnol. 2017, 182, 1065–1075. [Google Scholar] [CrossRef]








| Stage | Repeats | Temperature (°C) | Time |
|---|---|---|---|
| Stage1 Initial denaturation | Repeat 1 | 95 | 5 min |
| Stage2 Cyclic reaction | Repeats 40 | 95 | 10 s |
| 60 | 30 s | ||
| Stage3 Dissociation curve | Repeat 1 | 95 | 15 s |
| 60 | 1 min | ||
| Raise to 95 °C at 0.3 °C/s | 15 s |
| Gene | 5’-3’ Nucleotide Sequence | Amplification Product (bp) | Reference | Efficiency (%) |
|---|---|---|---|---|
| icaA | AAGCCAACGCACTCAATCAAGG | 151 | [27] | 73.92 |
| GGATTACCTGTAACCGCACCAAG | ||||
| icaD | ACCCAACGCTAAAATCATCG | 211 | [27] | 75.28 |
| GCGAAAATGCCCATAGTTTC | ||||
| icaR | TTTTCAGAGAAGGGGTATGACGG | 289 | This study | 72.75 |
| TTCCAGAAAATTCCTCAGGCGTA | ||||
| icaB | ATACCGGCGACTGGGTTTAT | 176 | This study | 75.06 |
| TTGCAAATCGTGGGTATGTGT | ||||
| icaC | CTTGGGTATTTGCACGCATT | 209 | [27] | 74.34 |
| GCAATATCATGCCGACACCT | ||||
| cflA | GCGTGGCTTCAGTGCTTGTA | 219 | This study | 70.08 |
| CCACACTCGTTTCGCCATTA | ||||
| fnbpA | CGACACAACCTCAAGACAATAGCGG | 146 | This study | 69.47 |
| CGTGGCTTACTTTCTGATGCCGTTC | ||||
| spa | GCTTAAAACCGCAAAATCACGC | 143 | This study | 68.26 |
| AACCTCAGGCACATTCAAAGC | ||||
| sasG | AACCTGGTGAAGAGCGAGTG | 209 | This study | 73.81 |
| GCTCGGCTTCTCTGGGTTTT | ||||
| 16S-rRNA | GGGACCCGCACAAGCGGTGG | 191 | [27] | 99.17 |
| GGGTTGCGCTCGTTGCGGGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, J.; Zheng, L.; Lu, Z.; Meng, F.; Bie, X. Research on the Biofilm Formation of Staphylococcus aureus after Cold Stress. Microorganisms 2021, 9, 1534. https://doi.org/10.3390/microorganisms9071534
Qiao J, Zheng L, Lu Z, Meng F, Bie X. Research on the Biofilm Formation of Staphylococcus aureus after Cold Stress. Microorganisms. 2021; 9(7):1534. https://doi.org/10.3390/microorganisms9071534
Chicago/Turabian StyleQiao, Jiaju, Liping Zheng, Zhaoxin Lu, Fanqiang Meng, and Xiaomei Bie. 2021. "Research on the Biofilm Formation of Staphylococcus aureus after Cold Stress" Microorganisms 9, no. 7: 1534. https://doi.org/10.3390/microorganisms9071534
APA StyleQiao, J., Zheng, L., Lu, Z., Meng, F., & Bie, X. (2021). Research on the Biofilm Formation of Staphylococcus aureus after Cold Stress. Microorganisms, 9(7), 1534. https://doi.org/10.3390/microorganisms9071534