Differentiation between Impacted and Unimpacted Microbial Communities of a Nitrogen Contaminated Aquifer
Abstract
:1. Introduction
1.1. Physicochemical Background
1.2. Microbial Background
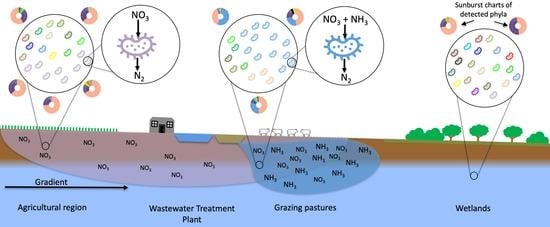
- Determine how nitrogen contamination influences the structure and ecosystem functions of the microbial communities of a nitrogen-contaminated aquifer.
- Use microbial community data and physicochemical data to explore how to differentiate between impacted and unimpacted microbial communities.
- Demonstrate the value of microbial functional group analysis in the characterisation of the impact and extent of groundwater contamination.
2. Materials and Methods
2.1. Study Area
2.2. Sampling Timeline and Locations
2.3. Sampling Techniques, Technology and Guidelines
2.4. Major Ions and Nutrients Analysis
2.5. Microbial Analysis
2.5.1. Sampling
2.5.2. DNA Extraction and Sequencing
2.5.3. File Preparation
2.5.4. Data Analysis
3. Results
3.1. Physicochemical Properties
3.2. Nitrogen and Physicochemical Properties in Contamination Zones
3.3. Nitrogen and Microbial Communities in Contamination Zones
3.4. Major Ecosystem Functions
3.4.1. Nitrogen Contaminated Bores (NCB)
- Bore DSE63273
- Bore RB23
- Bore RB10
- Bore RB06
- Bore RB07
3.4.2. Ammonia and Nitrogen Contaminated Bores (ANCB)
- Bore RB12
- Bore RB17
- Bore RB18
3.4.3. Uncontaminated Bores (UB)
- Bore BS-002
- Bore BS-004
4. Discussion
4.1. Major Ecosystem Function Analysis
4.2. Determining Microbial Background
4.3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Landrigan, P.J. The hidden costs of environmental contamination. Eur. Respir. J. 2012, 40, 286–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landrigan, P.J.; Fuller, R.; Hu, H.; Caravanos, J.; Cropper, M.L.; Hanrahan, D.; Sandilya, K.; Chiles, T.C.; Kumar, P.; Suk, W.A. Pollution and global health—An agenda for prevention. Environ. Health Perspect. 2018, 126, 084501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, A.M.; Steinberg, V.E. Health implications of nitrate and nitrite in drinking water: An update on methemoglobinemia occurrence and reproductive and developmental toxicity. Regul. Toxicol. Pharmacol. 1996, 23, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.H.; Patz, J.A. Reactive nitrogen and human health: Acute and long-term implications. Ambio A J. Hum. Environ. 2002, 31, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S. Chapter 2—Water quality worldwide. In Handbook of Water Purity and Quality, 2nd ed.; Ahuja, S., Ed.; Academic Press: Amsterdam, The Netherlands, 2021; pp. 19–33. [Google Scholar]
- Morrissy, J.G.; Currell, M.J.; Reichman, S.M.; Surapaneni, A.; Megharaj, M.; Crosbie, N.D.; Hirth, D.; Aquilina, S.; Rajendram, W.; Ball, A.S. Nitrogen contamination and bioremediation in groundwater and the environment: A review. Earth-Sci. Rev. 2021, 222, 103816. [Google Scholar] [CrossRef]
- Clark, C.M.; Bell, M.D.; Boyd, J.W.; Compton, J.E.; Davidson, E.A.; Davis, C.; Fenn, M.E.; Geiser, L.; Jones, L.; Blett, T.F. Nitrogen-Induced Terrestrial Eutrophication: Cascading Effects and Impacts on Ecosystem Services. Ecosphere 2017, 8, e01877. [Google Scholar] [CrossRef] [Green Version]
- Denk, T.R.; Mohn, J.; Decock, C.; Lewicka-Szczebak, D.; Harris, E.; Butterbach-Bahl, K.; Kiese, R.; Wolf, B. The nitrogen cycle: A review of isotope effects and isotope modeling approaches. Soil Biol. Biochem. 2017, 105, 121–137. [Google Scholar] [CrossRef] [Green Version]
- Katz, B.G. Nitrogen Overload: Environmental Degradation, Ramifications, and Economic Costs; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Arrigo, K.R. Marine microorganisms and global nutrient cycles. Nature 2005, 437, 349–355. [Google Scholar] [CrossRef]
- van der Meer, J.R. Environmental pollution promotes selection of microbial degradation pathways. Front. Ecol. Environ. 2006, 4, 35–42. [Google Scholar] [CrossRef]
- Reimann, C.; Garrett, R.G. Geochemical background—Concept and reality. Sci. Total Environ. 2005, 350, 12–27. [Google Scholar] [CrossRef]
- Panno, S.; Kelly, W.; Martinsek, A.; Hackley, K. Estimating background and threshold nitrate concentrations using probability graphs. Groundwater 2006, 44, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Steube, C.; Richter, S.; Griebler, C. First attempts towards an integrative concept for the ecological assessment of groundwater ecosystems. Hydrogeol. J. 2009, 17, 23–35. [Google Scholar] [CrossRef]
- Griebler, C.; Stein, H.; Kellermann, C.; Berkhoff, S.; Brielmann, H.; Schmidt, S.; Selesi, D.; Steube, C.; Fuchs, A.; Hahn, H.J. Ecological assessment of groundwater ecosystems—Vision or illusion? Ecol. Eng. 2010, 36, 1174–1190. [Google Scholar] [CrossRef]
- Morrissy, J.G.; Currell, M.J.; Reichman, S.M.; Surapaneni, A.; Megharaj, M.; Crosbie, N.D.; Hirth, D.; Aquilina, S.; Rajendram, W.; Ball, A.S. The variation in groundwater microbial communities in an unconfined aquifer contaminated by multiple nitrogen contamination sources. Water 2022, 14, 613. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Wang, H.; Cheng, Y.; Liu, H.; Jiang, Z.; Li, P.; Wang, Y. Effects of Different dissolved organic matter on microbial communities and arsenic mobilization in aquifers. J. Hazard. Mater. 2021, 411, 125146. [Google Scholar] [CrossRef]
- Guo, L.; Wang, G.; Sheng, Y.; Shi, Z.; Sun, X. Groundwater microbial communities and their connection to hydrochemical environment in golmud, Northwest China. Sci. Total Environ. 2019, 695, 133848. [Google Scholar] [CrossRef]
- Gilbert, D.; Amblard, C.; Bourdier, G.; Francez, A.J. Short-term effect of nitrogen enrichment on the microbial communities of a peatland. Hydrobiologia 1998, 373, 111–119. [Google Scholar] [CrossRef]
- Adebowale, T.; Surapaneni, A.; Faulkner, D.; McCance, W.; Wang, S.; Currell, M. Delineation of Contaminant sources and denitrification using isotopes of nitrate near a wastewater treatment plant in peri-urban settings. Sci. Total Environ. 2019, 651, 2701–2711. [Google Scholar] [CrossRef]
- McCance, W.; Jones, O.; Surapaneni, A.; Currell, M. Characterising sources of groundwater contamination: A guide to the use of novel groundwater tracers at wastewater treatment plants. Water E-J. 2020, 5, 1–16. [Google Scholar] [CrossRef]
- McCance, W.; Jones, O.A.H.; Cendón, D.I.; Edwards, M.; Surapaneni, A.; Chadalavada, S.; Wang, S.; Currell, M. Combining environmental isotopes with Contaminants of Emerging Concern (CECs) to characterise wastewater derived impacts on groundwater quality. Water Res. 2020, 182, 116036. [Google Scholar] [CrossRef]
- Leonard, J.G. Port Phillip Region Groundwater Systems-Future Use and Management. Department of Water Resources: Melbourne, VIC, Australia, 1992. [Google Scholar]
- EPA Victoria. Goundwater sampling guidelines. In Publication 669; State Government of Victoria: Melbourne, VIC, Australia, 2000. [Google Scholar]
- EPA Victoria. Sampling and analysis of waters, wastewaters, soils and wastes. In Publication IWRG701; State Government of Victoria: Melbourne, VIC, Australia, 2009. [Google Scholar]
- Koshlaf, E.; Shahsavari, E.; Aburto-Medina, A.; Taha, M.; Haleyur, N.; Makadia, T.H.; Morrison, P.D.; Ball, A.S. Bioremediation potential of diesel-contaminated Libyan soil. Ecotoxicol. Environ. Saf. 2016, 133, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16s rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adegoke, A.A.; Stenström, T.A.; Okoh, A.I. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Front. Microbiol. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; Van Der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T.; Boone, D.R.; Brenner, D.J.; De Vos, P.; Garrity, G.M.; Goodfellow, M.; Krieg, N.R.; Rainey, F.A.; Schleifer, K.H. Bergey’s Manual® of Systematic Bacteriology: Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria. Vol. 2; Springer Science & Business Media: Berlin, Germany, 2007. [Google Scholar]
- Yamada, T.; Sekiguchi, Y. Anaerolineaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 1–5. [Google Scholar]
- Soo, R.M.; Skennerton, C.T.; Sekiguchi, Y.; Imelfort, M.; Paech, S.J.; Dennis, P.G.; Steen, J.A.; Parks, D.H.; Tyson, G.W.; Hugenholtz, P. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol. Evol. 2014, 6, 1031–1045. [Google Scholar] [CrossRef] [Green Version]
- Willems, A.; Busse, J.; Goor, M.; Pot, B.; Falsen, E.; Jantzen, E.; Hoste, B.; Gillis, M.; Kersters, K.; Auling, G.; et al. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga Palleronii (formerly Pseudomonas Palleronii), Hydrogenophaga Pseudoflava (formerly Pseudomonas Pseudoflava and “Pseudomonas Carboxydoflava”), and Hydrogenophaga Taeniospiralis (formerly Pseudomonas taeniospiralis). Int. J. Syst. Evol. Microbiol. 1989, 39, 319–333. [Google Scholar]
- Hiraishi, A.; Ueda, Y. Rhodoplanes gen. nov., a new genus of phototrophic bacteria including Rhodopseudomonas Rosea as Rhodoplanes Roseus comb. nov. and Rhodoplanes Elegans Sp. Nov. Int. J. Syst. Evol. Microbiol. 1994, 44, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Imhoff, J.F.; Trüper, H.G.; Pfennig, N. Rearrangement of the Species and genera of the phototrophic “purple nonsulfur bacteria”. Int. J. Syst. Evol. Microbiol. 1984, 34, 340–343. [Google Scholar] [CrossRef] [Green Version]
- Dobbin, P.S.; Warren, L.H.; Cook, N.J.; McEwan, A.G.; Powell, A.K.; Richardson, D.J. Dissimilatory iron (III) reduction by Rhodobacter capsulatus. Microbiology 1996, 142, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Norris, P.R. Acidimicrobiales. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–2. [Google Scholar]
- Geissinger, O.; Herlemann, D.P.; Mörschel, E.; Maier, U.G.; Brune, A. The Ultramicrobacterium “Elusimicrobium minutum” Gen. Nov., Sp. Nov., the first cultivated representative of the termite group 1 phylum. Appl. Environ. Microbiol. 2009, 75, 2831–2840. [Google Scholar] [CrossRef] [Green Version]
- Kämpfer, P.; Lodders, N.; Falsen, E. Hydrotalea flava gen. nov., sp. nov., a new member of the phylum Bacteroidetes and allocation of the genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferruginibacter, Filimonas and Hydrotalea to the family Chitinophagaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 518–523. [Google Scholar] [PubMed]
- Spieck, E.; Eberhard, B. Nitrospira. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Qin, W.; Heal, K.R.; Ramdasi, R.; Kobelt, J.N.; Martens-Habbena, W.; Bertagnolli, A.D.; Amin, S.A.; Walker, C.B.; Urakawa, H.; Könneke, M.; et al. Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 2017, 67, 5067–5079. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Pseudomonadaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar]
- Washington, C.W., Jr. Legionella. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–44. [Google Scholar]
- Spröer, C.; Reichenbach, H.; Stackebrandt, E. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Evol. Microbiol. 1999, 49, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Tóth, E.; Szuróczki, S.; Kéki, Z.; Bóka, K.; Szili-Kovács, T.; Schumann, P. Gellertiella hungarica gen. nov., sp. nov., a novel bacterium of the family Rhizobiaceae isolated from a spa in Budapest. Int. J. Syst. Evol. Microbiol. 2017, 67, 4565–4571. [Google Scholar] [CrossRef]
- Pfennig, N.; Biebl, H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch. Microbiol. 1976, 110, 3–12. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Thermithiobacillaceae fam. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar]
- Kojima, H.; Shinohara, A.; Fukui, M. Sulfurifustis Variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 10, 3709–3713. [Google Scholar] [CrossRef]
- Umezawa, K.; Kojima, H.; Kato, Y.; Fukui, M. Dissulfurispira thermophila gen. nov., sp. nov., a thermophilic chemolithoautotroph growing by sulfur disproportionation, and proposal of novel taxa in the phylum Nitrospirota to reclassify the genus Thermodesulfovibrio. Syst. Appl. Microbiol. 2021, 44, 126184. [Google Scholar] [CrossRef]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 625–635. [Google Scholar]
- Taş, N.; Van Eekert, M.H.; De Vos, W.M.; Smidt, H. The little bacteria that can—Diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb. Biotechnol. 2010, 3, 389–402. [Google Scholar] [CrossRef] [Green Version]
- Reddy, G.S.; Garcia-Pichel, F. Description of Patulibacter americanus sp. nov., isolated from biological soil crusts, emended description of the genus Patulibacter Takahashi et al. 2006 and proposal of Solirubrobacterales ord. nov. and Thermoleophilales ord. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Jetten, M.S.; Op den Camp, H.J.; Kuenen, J.G.; Strous, M. “Candidatus brocadiaceae” fam. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–10. [Google Scholar]
- Moore, T.A.; Xing, Y.; Lazenby, B.; Lynch, M.D.; Schiff, S.; Robertson, W.D.; Timlin, R.; Lanza, S.; Ryan, M.C.; Aravena, R.; et al. Prevalence of anaerobic ammonium-oxidizing bacteria in contaminated groundwater. Environ. Sci. Technol. 2011, 45, 7217–7225. [Google Scholar] [CrossRef] [PubMed]
- Corteselli, E.M.; Aitken, M.D.; Singleton, D.R. Description of Immundisolibacter cernigliae gen. nov., sp. nov., a high-molecular-weight polycyclic aromatic hydrocarbon-degrading bacterium within the class Gammaproteobacteria, and proposal of Immundisolibacterales ord. nov. and Immundisolibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 925–931. [Google Scholar] [PubMed]
- Monchamp, M.E.; Spaak, P.; Pomati, F. Long term diversity and distribution of non-photosynthetic cyanobacteria in peri-alpine lakes. Front. Microbiol. 2019, 9, 3344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to cyanobacteria. elife 2013, 2, e01102. [Google Scholar] [CrossRef]
- Fukunaga, Y.; Kurahashi, M.; Sakiyama, Y.; Ohuchi, M.; Yokota, A.; Harayama, S. Phycisphaera Mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 2009, 55, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Vandamme, P.; Falsen, E.; Rossau, R.; Hoste, B.; Segers, P.; Tytgat, R.; De Ley, J. Revision of campylobacter, Helicobacter, and Wolinella taxonomy: Emendation of Generic Descriptions and Proposal of Arcobacter gen. nov. Int. J. Syst. Evol. Microbiol. 1991, 41, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Tarrand, J.J.; Krieg, N.R.; Döbereiner, J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can. J. Microbiol. 1978, 24, 967–980. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Zhang, W.; Li, X.; Zhou, Y.; Li, D.; Wang, Y.; Tian, J.; Jiang, W.; Zhang, Z.; et al. Physiological characteristics of magnetospirillum gryphiswaldense msr-1 that control cell growth under high-iron and low-oxygen conditions. Sci. Rep. 2017, 7, 2800. [Google Scholar] [CrossRef] [Green Version]
- Kojima, H.; Fukui, M. Sulfuritalea hydrogenivorans gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2011, 61, 1651–1655. [Google Scholar] [CrossRef] [Green Version]
- Balk, M.; Altınbaş, M.; Rijpstra, W.I.C.; Damste, J.S.S.; Stams, A.J. Desulfatirhabdium butyrativorans gen. nov., sp. nov., a butyrate-oxidizing, sulfate-reducing bacterium isolated from an anaerobic bioreactor. Int. J. Syst. Evol. Microbiol. 2008, 58, 110–115. [Google Scholar] [CrossRef]
- Cummings, D.E.; Caccavo Jr, F.; Spring, S.; Rosenzweig, R.F. Ferribacterium limneticum, gen. nov., sp. nov., an Fe (III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch. Microbiol. 1999, 171, 183–188. [Google Scholar] [CrossRef]
- Bowman, J.P.; Sly, L.I.; Nichols, P.D.; Hayward, A.C. Revised Taxonomy of the methanotrophs: Description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Evol. Microbiol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Ferry, J.G.; Smith, P.H.; Wolfe, R.S. Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii Sp.Nov. Int. J. Syst. Evol. Microbiol. 1974, 24, 465–469. [Google Scholar] [CrossRef]
- Kodama, Y.; Watanabe, K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2297–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, H.; Fukui, M. Sulfuricella denitrificans gen. nov., sp. nov., a sulfur-oxidizing autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2010, 60, 2862–2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yordy, J.R.; Weaver, T.L. Methylobacillus: A new genus of obligately methylotrophic bacteria. Int. J. Syst. Evol. Microbiol. 1977, 27, 247–255. [Google Scholar] [CrossRef]
- Higashioka, Y.; Kojima, H.; Watanabe, M.; Fukui, M. Desulfatitalea tepidiphila gen. nov., sp. nov., a sulfate-reducing bacterium isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 761–765. [Google Scholar] [CrossRef] [Green Version]
- Kizina, J. Insights into the Biology of Candidate Division Op3 Lim Populations; Universität Bremen: Bremen, Germany, 2017. [Google Scholar]
- Saidi-Mehrabad, A.; He, Z.; Tamas, I.; Sharp, C.E.; Brady, A.L.; Rochman, F.F.; Bodrossy, L.; Abell, G.C.; Penner, T.; Dong, X.; et al. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013, 7, 908–921. [Google Scholar] [CrossRef] [Green Version]
- De Silva, S.; Ball, A.S.; Shahsavari, E.; Indrapala, D.V.; Reichman, S.M. The effects of vehicular emissions on the activity and diversity of the roadside soil microbial community. Environ. Pollut. 2021, 277, 116744. [Google Scholar] [CrossRef]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Pseudomonas pickettii: A common soil and groundwater aerobic bacteria with pathogenic and biodegradation properties. Ecotoxicol. Environ. Saf. 2000, 47, 105–111. [Google Scholar] [CrossRef]




| Min-max Physiochemical Properties | DSE63273 | RB23 | RB10 | RB12 | RB17 | RB18 | RB06 | RB07 | BS-002 | BS-004 |
|---|---|---|---|---|---|---|---|---|---|---|
| Dissolved Oxygen | 4.47 | 4.77–5.92 | 0.22–0.51 | 0.15–0.47 | 0.2–0.64 | 0.17–0.56 | 0.12–0.61 | 0.22–0.76 | 0.24–0.74 | 0.17–0.69 |
| EC (field) (uS/cm) | 2945 | 1434–1487 | 2357–2659 | 2192–2641 | 1486–1723 | 1309–1480 | 1915–2633 | 3342–3503 | 940–1004 | 718–776 |
| pH (Field) (unitless) | 6.70 | 6.93–7.27 | 6.68–6.73 | 6.69–6.81 | 6.84–7.01 | 6.87–7.24 | 6.79–7.19 | 6.73–6.78 | 7.37–7.43 | 7.44–7.77 |
| Redox Potential (Field) (mV) | 107.0 | 28.9–111.3 | 21.1–61.2 | 23.3–68.7 | 29.1–77.2 | 20.7–79.9 | 33.4–35.9 | −8.4–24.0 | −59.6–5.5 | −106.9–−83.5 |
| Alkalinity (total) as CaCO3 | 315 | 170–270 | 430–610 | 450–560 | 350–370 | 350–370 | 270–330 | 410–420 | 240–250 | 210–210 |
| Bicarbonate, as Bicarbonate | 385 | 210–329.4 | 530–744.2 | 550–683.2 | 430–450 | 430–450 | 330–400 | 490–510 | 300–300 | 250–260 |
| Sulfate as SO42− | 410 | 24–45 | 580–760 | 330–530 | 220–330 | 130–190 | 490–1100 | 1500–1500 | 7–8 | 26–34 |
| Anionic Strength (meq/L) | 32 | 13–17 | 29–34 | 26–29 | 17–20 | 14–16 | 22–37 | 48–48 | 9–10 | 7–8 |
| Cationic Strength (meq/L) | 28.5 | 12–14 | 27–34 | 24–31 | 18–18 | 13–15 | 18–37 | 36–36 | 8–9 | 7–7 |
| Calcium | 390 | 150–170 | 350–430 | 230–320 | 200–200 | 130–170 | 210–480 | 470–570 | 83–84 | 47–51 |
| Magnesium | 41 | 15–19 | 50–69 | 48–61 | 27–32 | 22–25 | 34–63 | 70–81 | 11–11 | 25–28 |
| Sodium | 135 | 75–85 | 110–140 | 120–160 | 120–130 | 100–110 | 110–170 | 160–210 | 78–82 | 49–52 |
| Potassium | 1.2 | 1–1 | 11–14 | 31–40 | 0.8–0.8 | 3–3.5 | 0.8–0.8 | 1–7 | 1–1.3 | 4–4 |
| Chloride | 230 | 150–170 | 170–210 | 170–210 | 150–160 | 110–140 | 180–250 | 300–320 | 160–170 | 93–100 |
| Bromide | 1600 | 680–970 | 3700–4700 | 1200–1400 | 530–830 | 510–600 | 700–940 | 1500–1600 | 380–450 | 230–280 |
| Ammonia as N | 0.15 | <0.1–<0.1 | <0.1–<0.1 | 5.5–26 | <0.1–<0.1 | 3.3–4.6 | <0.1–<0.1 | 0.1–0.1 | 0.1–0.1 | 0.2–0.2 |
| Nitrate (as N) | 150 | 64–83 | 14–54 | 40–63 | 12–15 | 11–18 | 7.5–18 | 0.22–0.22 | 0.01–0.01 | <0.01–<0.01 |
| Nitrogen (Total Oxidised) | 155 | 64–88 | 15–54 | 40–63 | 12–15 | 11–15 | 7.15–18 | 0.26–0.26 | 0.01–0.01 | <0.01–<0.01 |
| Nitrogen (Total) | 155 | 65–88 | 15–55 | 60–76 | 13–16 | 18–20 | 8.2–20 | 0.9–1.2 | 0.07–0.2 | 0.3–0.66 |
| TOC | 3.7 | 1.3–2.5 | 10–13 | 10–12 | 1.6–7.1 | 1.3–6.1 | 4.4–8.1 | 13–16 | 2.6–2.6 | 2.7–3.1 |
| Organic Nitrogen, as N | 0.6 | 0.2–0.4 | 0.8–1.2 | 0.2–0.2 | 0.4–0.6 | 0.1–0.1 | 0.7–0.8 | 0.7–1.1 | 0.1–0.2 | 0.1–0.4 |
| Kjeldahl Nitrogen Total | 3.2 | 0.2–0.7 | 0.8–1.3 | 5.5–26 | 0.5–2 | 3.1–8.3 | 0.8–2.2 | 0.9–0.94 | 0.06–0.2 | 0.3–0.46 |
| Phosphorus | 0.009 | <0.005–<0.005 | <0.005–<0.005 | 0.05–0.05 | <0.005–<0.005 | 0.006–0.006 | <0.005–<0.005 | 0.08–0.08 | <0.005–<0.005 | 0.05–0.05 |
| TDS | 2250 | 960–1200 | 1700–1900 | 1400–1600 | 1000–1200 | 790–840 | 1200–2100 | 2700–2800 | 440–480 | 300–360 |
| Iron (Filtered) | 0.02 | 0.003–0.003 | 0.015–0.015 | 0.009–0.009 | 0.003–0.003 | 0.003–0.003 | <0.1–<0.1 | 0.9–1.2 | 0.096–0.13 | 0.57–1.4 |
| Molybdenum (Filtered) | <0.001 | 0.0006–0.0006 | <0.001–<0.001 | <0.001–<0.001 | 0.0002–0.0002 | 0.0001–0.0001 | <0.001–<0.001 | <0.001–<0.001 | 0.0002–0.0002 | 0.004–0.0055 |
| Nickel (Filtered) | <0.001 | 0.0011–0.0011 | 0.004–0.011 | 0.004–0.0054 | 0.00078–0.003 | 0.00059–0.00059 | 0.002–0.002 | 0.001–0.002 | 0.002–0.0051 | 0.00018–0.00018 |
| Zinc (Filtered) | 0.0295 | 0.006–0.014 | 0.01–0.013 | 0.0086–0.015 | 0.0043–0.011 | 0.004–0.007 | 0.005–0.032 | 0.01–0.01 | 0.0026–0.008 | 0.0007–0.003 |
| Bore Depth (CS) (m) | 25.1 | 11.1 | 5.3 | 5.1 | 4.7 | 10.8 | 9.1 | 4.6 | 26.4 | 5.5 |
| Number of samples | 1 | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| MANOVA (p) | PERMANOVA (Pr(>F)) | ||
|---|---|---|---|
| Analysis Groups | All Nitrogen Variables | Contamination Zones | Ammonia (NH4+) |
| all | 0.0052 | 0.0002 | 0.0012 |
| UB-ANCB | 0.0438 | 0.0099 | 0.0453 |
| UB-NCB | 0.0079 | 0.4048 | 0.4323 |
| NCB-ANCB | 0.0133 | 0.0001 | 0.0006 |
| Min Max Microbial Community Descriptors | DSE63273 | RB23 | RB10 | RB12 | RB17 | RB18 | RB06 | RB07 | BS-002 | BS-004 |
| Non-Chimeric Sequences | 6403 | 12,150–26,674 | 1584–21,782 | 7353–23,760 | 1542–21,920 | 1691–30,838 | 1964–2284 | 15,628–36,019 | 9588–33,592 | 1809–13,032 |
| OTUs | 1289 | 1153–1263 | 263–2590 | 1705–2391 | 415–1643 | 398–1591 | 556–758 | 2894–2906 | 940–1534 | 379–1882 |
| Total OTUs | 1289 | 1725 | 2926 | 3055 | 2276 | 2051 | 1035 | 3711 | 1682 | 2146 |
| ACE | 1429 | 1227–1309 | 291–2717 | 1907–2551 | 432–1790 | 409–1703 | 578–837 | 3075–3099 | 994–1589 | 389–2066 |
| Chao1 estimated no species | 1524 | 1260–1404 | 779–3333 | 2069–5870 | 574–1772 | 593–1916 | 858–1664 | 3345–6421 | 1039–1614 | 610–2090 |
| Gini-Simpson | 0.01 | 0.01–0.04 | 0.01–0.11 | 0.04–0.13 | 0.02–0.1 | 0.02–0.1 | 0.01–0.01 | 0.01–0.01 | 0.03–0.05 | 0.01–0.02 |
| Shannon rare | 5.41 | 4.6–5.72 | 3.56–6.58 | 3.58–5.35 | 3.98–4.75 | 3.92–4.91 | 5.19–5.55 | 6.34–6.45 | 4.65–4.92 | 4.87–6.03 |
| Pielou’s Evenness | 0.76 | 0.64–0.81 | 0.64–0.84 | 0.48–0.7 | 0.54–0.79 | 0.54–0.82 | 0.82–0.84 | 0.8–0.81 | 0.63–0.72 | 0.8–0.82 |
| Percent of OTUs in Common | DSE63273 | RB23 | RB10 | RB12 | RB17 | RB18 | RB06 | RB07 | BS-002 | BS-004 |
| DSE63273 | 12.21 | 16.47 | 16.77 | 14.08 | 13.72 | 12.22 | 23.12 | 17.52 | 22.46 | |
| RB23 | 12.21 | 24.26 | 22.03 | 27.54 | 34.14 | 26.14 | 21.69 | 33.82 | 20.25 | |
| RB10 | 16.47 | 24.26 | 52.62 | 40.48 | 32.30 | 20.65 | 41.54 | 17.16 | 23.83 | |
| RB12 | 16.77 | 22.03 | 52.62 | 42.24 | 36.63 | 19.24 | 42.95 | 16.16 | 23.98 | |
| RB17 | 14.08 | 27.54 | 40.48 | 42.24 | 49.83 | 24.05 | 31.35 | 18.29 | 21.72 | |
| RB18 | 13.72 | 34.14 | 32.30 | 36.63 | 49.83 | 26.11 | 27.37 | 25.73 | 21.16 | |
| RB06 | 12.22 | 26.14 | 20.65 | 19.24 | 24.05 | 26.11 | 18.03 | 20.81 | 17.25 | |
| RB07 | 23.12 | 21.69 | 41.54 | 42.95 | 31.35 | 27.37 | 18.03 | 21.60 | 38.96 | |
| BS-002 | 17.52 | 33.82 | 17.16 | 16.16 | 18.29 | 25.73 | 20.81 | 21.60 | 23.64 | |
| BS-004 | 22.46 | 20.25 | 23.83 | 23.98 | 21.72 | 21.16 | 17.25 | 38.96 | 23.64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morrissy, J.G.; Reichman, S.M.; Currell, M.J.; Surapaneni, A.; Megharaj, M.; Crosbie, N.D.; Hirth, D.; Aquilina, S.; Rajendram, W.; Ball, A.S. Differentiation between Impacted and Unimpacted Microbial Communities of a Nitrogen Contaminated Aquifer. Environments 2022, 9, 128. https://doi.org/10.3390/environments9100128
Morrissy JG, Reichman SM, Currell MJ, Surapaneni A, Megharaj M, Crosbie ND, Hirth D, Aquilina S, Rajendram W, Ball AS. Differentiation between Impacted and Unimpacted Microbial Communities of a Nitrogen Contaminated Aquifer. Environments. 2022; 9(10):128. https://doi.org/10.3390/environments9100128
Chicago/Turabian StyleMorrissy, Justin G., Suzie M. Reichman, Matthew J. Currell, Aravind Surapaneni, Mallavarapu Megharaj, Nicholas D. Crosbie, Daniel Hirth, Simon Aquilina, William Rajendram, and Andrew S. Ball. 2022. "Differentiation between Impacted and Unimpacted Microbial Communities of a Nitrogen Contaminated Aquifer" Environments 9, no. 10: 128. https://doi.org/10.3390/environments9100128
APA StyleMorrissy, J. G., Reichman, S. M., Currell, M. J., Surapaneni, A., Megharaj, M., Crosbie, N. D., Hirth, D., Aquilina, S., Rajendram, W., & Ball, A. S. (2022). Differentiation between Impacted and Unimpacted Microbial Communities of a Nitrogen Contaminated Aquifer. Environments, 9(10), 128. https://doi.org/10.3390/environments9100128