Convolutional Neural Networks for Breast Density Classification: Performance and Explanation Insights
Abstract
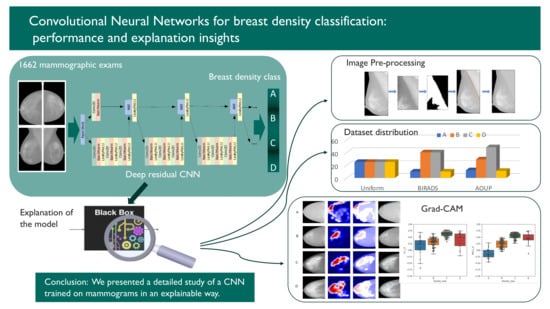
:1. Introduction
2. Materials and Methods
2.1. Data
- All exam reports were required to be negative. Whenever possible, a later mammographic exam in medical records has been examined to verify the current state of health of women.
- Badly exposed X-ray mammograms were not collected.
- Only exams including all the four projections usually acquired in mammography (cranio-caudal—CC—and medio-lateral oblique—MLO—of left and right breast) were chosen.
2.2. Methods
- The different proportion of mammograms belonging to the four density categories in the training and test sets;
- Either including or not an image pre-processing step.
2.2.1. Data Preparation and Pre-Processing
2.2.2. Standard Image Pre-Processing Step
2.2.3. Additional Pre-Processing Step: Pectoral Muscle Removal
2.2.4. Data Augmentation for CNN Training
- Random zoom in a range of 0.2;
- Width shift in a range of 0.2 of the whole input image;
- Height shift in a range of 0.2 of the whole input image;
- Random rotations with a range of 10 degrees.
2.2.5. Classifier Training
- Forty-one convolutional layers organized in 12 similar blocks;
- Training performed in batches of four images;
- Loss function: Categorical Cross-Entropy;
- Optimizer: Stochastic Gradient Descent (SGD);
- Regularization: Batch Normalization;
- Learning rate = 0.1, Decay = 0.1, Patience = 15, Monitor = validation loss.
2.2.6. Model Explanation
2.2.7. Evaluation of the Explanation Framework
3. Results
3.1. Evaluation of the Effect of Sample Composition on CNN Training
3.2. Implementation and Visual Assessment of the Grad-CAM Technique
3.3. Evaluation of the Impact of Pectoral Muscle Removal
3.4. Quantitative Evaluation of the Explanation Framework
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FFDM | Full Field Digital Mammography |
| ACR | American College of Radiology |
| BI-RADS | Breast Imaging Reporting and Data Systems |
| SVM | Support Vector Machine |
| CNN | Convolutional Neural Network |
| grad-CAM | grad Class Activation Map |
| RADIOMA | Ionizing Radiation in MAmmography |
| AOUP | Azienda Ospedaliero Universitaria Pisana |
| CC | Cranio-Caudal (projection) |
| MLO | Medio Lateral-Oblique (projection) |
| DICOM | Digital Imaging and COmmunication in Medicine |
| PNG | Portable Network Graphics |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019: Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- Løberg, M.; Lousdal, M.L.; Bretthauer, M.; Kalager, M. Benefits and harms of mammography screening. Breast Cancer Res. 2015, 17, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dance, D.R.; Christofides, S.; McLean, I.; Maidment, A.; Ng, K. Diagnostic Radiology Physics; Non-Serial Publications, International Atomic Energy Agency: Vienna, Austria, 2014. [Google Scholar]
- The Independent UK Panel on Breast Cancer Screening; Marmot, M.G.; Altman, D.G.; Cameron, D.A.; Dewar, J.A.; Thompson, S.G.; Wilcox, M. The benefits and harms of breast cancer screening: An independent review: A report jointly commissioned by Cancer Research UK and the Department of Health (England) October 2012. Br. J. Cancer 2013, 108, 2205–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Orsi, C. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Miglioretti, D.L.; Lange, J.; van den Broek, J.J.; Lee, C.I.; van Ravesteyn, N.T.; Ritley, D.; Kerlikowske, K.; Fenton, J.J.; Melnikow, J.; de Koning, H.J.; et al. Radiation-Induced Breast Cancer Incidence and Mortality From Digital Mammography Screening: A Modeling Study. Ann. Intern. Med. 2016, 164, 205. [Google Scholar] [CrossRef]
- McCormack, V.A. Breast Density and Parenchymal Patterns as Markers of Breast Cancer Risk: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, N.F.; Byng, J.W.; Jong, R.A.; Fishell, E.K.; Little, L.E.; Miller, A.B.; Lockwood, G.A.; Tritchler, D.L.; Yaffe, M.J. Quantitative Classification of Mammographic Densities and Breast Cancer Risk: Results From the Canadian National Breast Screening Study. JNCI J. Natl. Cancer Inst. 1995, 87, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Tyrer, J.; Duffy, S.W.; Cuzick, J. A breast cancer prediction model incorporating familial and personal risk factors. Stat. Med. 2004, 23, 1111–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciatto, S.; Houssami, N.; Apruzzese, A.; Bassetti, E.; Brancato, B.; Carozzi, F.; Catarzi, S.; Lamberini, M.; Marcelli, G.; Pellizzoni, R.; et al. Categorizing breast mammographic density: Intra- and interobserver reproducibility of BI-RADS density categories. Breast 2005, 14, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Kumar, I.; Bhadauria, H.S.; Virmani, J.; Thakur, S. A classification framework for prediction of breast density using an ensemble of neural network classifiers. Biocybern. Biomed. Eng. 2017, 37, 217–228. [Google Scholar] [CrossRef]
- Bovis, K.; Singh, S. Classification of mammographic breast density using a combined classifier paradigm. Med. Image Underst. Anal. 2002, pp. 1–4. Available online: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.19.1806&rep=rep1&type=pdf (accessed on 1 November 2021).
- Oliver, A.; Freixenet, J.; Zwiggelaar, R. Automatic Classification of Breast Density. In Proceedings of the IEEE International Conference on Image Processing 2005, Genova, Italy, 14 September 2005; pp. 1258–1261. [Google Scholar]
- Oliver, A.; Freixenet, J.; Marti, R.; Pont, J.; Perez, E.; Denton, E.; Zwiggelaar, R. A Novel Breast Tissue Density Classification Methodology. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzikopoulos, S.D.; Mavroforakis, M.E.; Georgiou, H.V.; Dimitropoulos, N.; Theodoridis, S. A fully automated scheme for mammographic segmentation and classification based on breast density and asymmetry. Comput. Methods Programs Biomed. 2011, 102, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Petroudi, S.; Kadir, T.; Brady, M. Automatic classification of mammographic parenchymal patterns: A statistical approach. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat. No.03CH37439), Cancun, Mexico, 17–21 September 2003; pp. 798–801. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kortesniemi, M.; Tsapaki, V.; Trianni, A.; Russo, P.; Maas, A.; Källman, H.E.; Brambilla, M.; Damilakis, J. The European Federation of Organisations for Medical Physics (EFOMP) White Paper: Big data and deep learning in medical imaging and in relation to medical physics profession. Phys. Med. 2018, 56, 90–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. arXiv 2016, arXiv:1602.04938. [Google Scholar]
- Suckling, J.; Parker, J.; Dance, D.; Astley, S.; Hutt, I.; Boggis, C.; Ricketts, I.; Stamatakis, E.; Cerneaz, N.; Kok, S.; et al. Mammographic Image Analysis Society (MIAS) Database v1.21; 2015. Available online: https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 21 December 2021).
- Lizzi, F.; Laruina, F.; Oliva, P.; Retico, A.; Fantacci, M.E. Residual Convolutional Neural Networks to Automatically Extract Significant Breast Density Features. In Computer Analysis of Images and Patterns; Vento, M., Percannella, G., Colantonio, S., Giorgi, D., Matuszewski, B.J., Kerdegari, H., Razaak, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 28–35. [Google Scholar]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef]
- Sottocornola, C.; Traino, A.; Barca, P.; Aringhieri, G.; Marini, C.; Retico, A.; Caramella, D.; Fantacci, M.E. Evaluation of Dosimetric Properties in Full Field Digital Mammography (FFDM)-Development of a New Dose Index. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies-Volume 1, Madeira, Portugal, 19–21 January 2018; SciTePress: Setúbal, Portugal, 2018; pp. 212–217. [Google Scholar] [CrossRef]
- Wenger, R. Isosurfaces: Geometry, Topology, and Algorithms; CRC Press: London, UK, 2013; Chapter 2; pp. 17–44. [Google Scholar]
- Maple, C. Geometric design and space planning using the marching squares and marching cube algorithms. In Proceedings of the 2003 International Conference on Geometric Modeling and Graphics, London, UK, 16–18 July 2003; pp. 90–95. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 21 December 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple Comparisons Using Rank Sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Lehman, C.D.; Yala, A.; Schuster, T.; Dontchos, B.; Bahl, M.; Swanson, K.; Barzilay, R. Mammographic Breast Density Assessment Using Deep Learning: Clinical Implementation. Radiology 2019, 290, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Gandomkar, Z.; Suleiman, M.E.; Demchig, D.; Brennan, P.C.; McEntee, M.F. BI-RADS density categorization using deep neural networks. Proc. SPIE 2019, 10952, 109520N. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Berg, W.A.; Peng, H.; Luo, Y.; Jankowitz, R.C.; Wu, S. A deep learning method for classifying mammographic breast density categories. Med. Phys. 2018, 45, 314–321. [Google Scholar] [CrossRef]
- Saffari, N.; Rashwan, H.A.; Abdel-Nasser, M.; Singh, V.K.; Arenas, M.; Mangina, E.; Herrera, B.; Puig, D. Fully automated breast density segmentation and classification using deep learning. Diagnostics 2020, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Weiss, G.M.; Provost, F. Learning When Training Data are Costly: The Effect of Class Distribution on Tree Induction. J. Artif. Intell. Res. 2003, 19, 315–354. [Google Scholar] [CrossRef] [Green Version]







| A | B | C | D | |
|---|---|---|---|---|
| N. of exams | 200 | 473 | 804 | 185 |
| Average age (years) | 61 | 57 | 51 | 46 |
| AOUP | BIRADS | Uniform | ||
|---|---|---|---|---|
| Test Set | Test Set | Test Set | ||
| BIRADS Training set | test accuracy (%) | 79.1 | 83.1 | 73.6 |
| recall (%) | 75.2 | 80.1 | 73.6 | |
| precision (%) | 82.6 | 87.9 | 79.0 | |
| AOUP Training set | test accuracy (%) | 78.5 | 79.7 | 73.6 |
| recall (%) | 74.2 | 77.9 | 73.6 | |
| precision (%) | 81.2 | 83.0 | 79.4 | |
| Uniform Training set | test accuracy (%) | 72.8 | 72.9 | 77.8 |
| recall (%) | 78.9 | 79.9 | 77.8 | |
| precision (%) | 69.5 | 68.8 | 78.0 |
| Precision | Recall | Accuracy | |
|---|---|---|---|
| with PM | 81.1% | 78.1% | 79.9% |
| without PM | 83.3% | 80.3% | 82.0% |
| A | B | C | D | |
|---|---|---|---|---|
| A | 1 | p = 0.43 | p < 0.05 | p = 0.12 |
| B | p = 0.43 | 1 | p < 0.05 | p = 0.16 |
| C | p < 0.05 | p < 0.05 | 1 | p < 0.05 |
| D | p = 0.12 | 0.16 | p < 0.05 | 1 |
| A | B | C | D | |
| A | 1 | p < 0.05 | p < 0.05 | p < 0.05 |
| B | p < 0.05 | 1 | p < 0.05 | p < 0.05 |
| C | p < 0.05 | p < 0.05 | 1 | p = 0.20 |
| D | p < 0.05 | p < 0.05 | p = 0.20 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lizzi, F.; Scapicchio, C.; Laruina, F.; Retico, A.; Fantacci, M.E. Convolutional Neural Networks for Breast Density Classification: Performance and Explanation Insights. Appl. Sci. 2022, 12, 148. https://doi.org/10.3390/app12010148
Lizzi F, Scapicchio C, Laruina F, Retico A, Fantacci ME. Convolutional Neural Networks for Breast Density Classification: Performance and Explanation Insights. Applied Sciences. 2022; 12(1):148. https://doi.org/10.3390/app12010148
Chicago/Turabian StyleLizzi, Francesca, Camilla Scapicchio, Francesco Laruina, Alessandra Retico, and Maria Evelina Fantacci. 2022. "Convolutional Neural Networks for Breast Density Classification: Performance and Explanation Insights" Applied Sciences 12, no. 1: 148. https://doi.org/10.3390/app12010148
APA StyleLizzi, F., Scapicchio, C., Laruina, F., Retico, A., & Fantacci, M. E. (2022). Convolutional Neural Networks for Breast Density Classification: Performance and Explanation Insights. Applied Sciences, 12(1), 148. https://doi.org/10.3390/app12010148