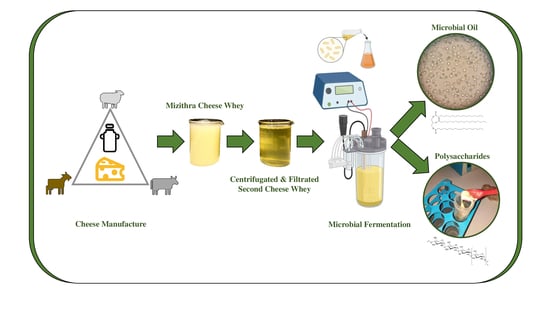
Biotechnological Conversions of Mizithra Second Cheese Whey by Wild-Type Non-Conventional Yeast Strains: Production of Yeast Cell Biomass, Single-Cell Oil and Polysaccharides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Media
2.3. Analytical Methods
2.4. Data Analysis
3. Results
3.1. Trials on Lactose-Based Media
3.2. Trials in Treated Second Cheese Whey
3.3. Polysaccharides by P. laurentii NRRL Y-2536
3.4. Enhanced Lipid Production through Cultivation of C. curvatus ATCC 20509 in Fed-Batch Conditions
3.5. Yeast Lipid Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaza, S.; Yao, L.; Bhada-Tata, P.; Van Woerden, F. What a Waste 2.0: A Global Snapshot of Solid Waste Management to 2050. In Urban Development; World Bank: Washington, DC, USA, 2018. [Google Scholar]
- Coma, M.; Chatzifragkou, A. Chemicals from food supply chain by-products and waste streams. Molecules 2019, 24, 978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papanikolaou, S.; Aggelis, G. Microbial products from wastes and residues. FEMS Microbiol. Lett. 2020, 367, fnaa156. [Google Scholar] [CrossRef] [PubMed]
- Lappa, I.K.; Papadaki, A.; Kachrimanidou, V.; Terpou, A.; Koulougliotis, D.; Eriotou, E.; Kopsahelis, N. Cheese whey processing: Integrated biorefinery concepts and emerging food applications. Foods 2019, 8, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, M.-Y.; Mourti, C.; Tatoulis, T.; Akratos, C.S.; Tekerlekopoulou, A.G.V.; Ayenas, D.V. Effect of hydraulic retention time, temperature, and organic load on a horizontal subsurface flow constructed wetland treating cheese whey wastewater. J. Chem. Technol. Biotechnol. 2016, 91, 726–732. [Google Scholar] [CrossRef]
- Panesar, P.S.; Kennedy, J.F. Biotechnological approaches for the value addition of whey. Crit. Rev. Biotechnol. 2011, 32, 327–348. [Google Scholar] [CrossRef] [PubMed]
- Eurostat. Milk and Milk Product Statistics. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Milk_and_milk_product_statistics#Milk_production (accessed on 10 August 2020).
- Georgala, A.; Kaminarides, S. A review of Greek whey cheeses: A description of their characteristics. In Proceedings of the 10th Panhellenic Congress of Agricultural Engineering, Athens, Greece, 10–13 September 2017. [Google Scholar]
- Chatzipaschali, A.A.; Stamatis, A.G. Biotechnological utilization with a focus on anaerobic treatment of cheese whey: Current status and prospects. Energies 2012, 5, 3492–3525. [Google Scholar] [CrossRef] [Green Version]
- Farizoglu, B.; Keskinler, B.; Yildiz, E.; Nuhoglu, A. Cheese whey treatment performance of an aerobic jet loop membrane bioreactor. Proc. Biochem. 2004, 39, 2283–2291. [Google Scholar] [CrossRef]
- Antonopoulou, G.; Stamatelatou, K.; Venetsaneas, N.; Kornaros, M.; Lyberatos, G. Biohydrogen and methane production from cheese whey in a two-stage anaerobic process. Ind. Eng. Chem. Res. 2008, 47, 5227–5233. [Google Scholar] [CrossRef]
- Rico, C.; Muñoz, N.; Rico, J.L. Anaerobic co-digestion of cheese whey and the screened liquid fraction of dairy manure in a single continuously stirred tank reactor process: Limits in co-substrate ratios and organic loading rate. Bioresour. Technol. 2015, 189, 327–333. [Google Scholar] [CrossRef]
- Shouvik, S.; Bikram, B.; Jae-Hoon, H.; El-Sayed, S.; Pradip, K.C.; Byong-Hun, J. Microbial symbiosis: A network towards biomethanation. Trends Microbiol. 2020, 28, 968–984. [Google Scholar]
- Venetsaneas, N.; Antonopoulou, G.; Stamatelatou, K.; Kornaros, M.; Lyberatos, G. Using cheese whey for hydrogen and methane generation in a two-stage continuous process with alternative pH controlling approaches. Bioresour. Technol. 2009, 100, 3713–3717. [Google Scholar] [CrossRef] [PubMed]
- Asunis, F.; De Gioannis, G.; Isipato, M.; Muntoni, A.; Polettini, A.; Pomi, R.; Rossi, A.; Spiga, D. Control of fermentation duration and pH to orient biochemicals and biofuels production from cheese whey. Bioresour. Technol. 2019, 289, 121722. [Google Scholar] [CrossRef] [PubMed]
- Schingoethe, D.J. Whey utilization in animal feeding: A summary and evaluation. J. Dairy Sci. 1976, 59, 556–570. [Google Scholar] [CrossRef]
- Early, R. Dairy products and milk-based food ingredients. Nat. Food Addit. Ingred. Flavour. 2012, 17, 417–445. [Google Scholar]
- Kilara, A.; Vaghela, M.N. Whey proteins. Protein Food Proc. 2018, 4, 93–126. [Google Scholar]
- Vamvakaki, A.-N.; Kandarakis, I.; Kaminarides, S.; Komaitis, M.; Papanikolaou, S. Cheese whey as a renewable substrate for microbial lipid and biomass production by Zygomycetes. Eng. Life Sci. 2010, 10, 348–360. [Google Scholar] [CrossRef]
- Yadav, J.S.S.; Bezawada, J.; Ajila, C.M.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Mixed culture of Kluyveromyces marxianus and Candida krusei for single-cell protein production and organic load removal from whey. Bioresour. Technol. 2014, 164, 119–127. [Google Scholar] [CrossRef]
- Yadav, J.S.S.; Yan, S.; Ajila, C.M.; Bezawada, J.; Tyagi, R.D.; Surampalli, R.Y. Food-grade single-cell protein production, characterization and ultrafiltration recovery of residual fermented whey proteins from whey. Food Bioprod. Processing 2016, 99, 156–165. [Google Scholar] [CrossRef]
- Aggelopoulos, T.; Katsieris, K.; Bekatorou, A.; Pandey, A.; Banat, I.M.; Koutinas, A.A. Solid state fermentation of food waste mixtures for single cell protein, aroma volatiles and fat production. Food Chem. 2014, 145, 710–716. [Google Scholar] [CrossRef]
- Papadaki, A.; Lappa, I.K.; Kachrimanidou, V.; Gonou-Zagou, Z.; Kopsahelis, N. Trametes versicolor as a natural source of bioactive compounds for the production of whey protein films with functional properties: A holistic approach to valorize cheese whey. Waste Biom. Valor. 2022, in press. [CrossRef]
- Takakuwa, N.; Saito, K. Conversion of beet molasses and cheese whey into fatty acid methyl esters by the yeast Cryptococcus curvatus. J. Oleo Sci. 2010, 59, 255–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanha, R.F.; Mariano, A.P.; de Morais, L.A.; Scramin, S.; Monteiro, R.T. Optimization of lipids production by Cryptococcus laurentii 11 using cheese whey with molasses. Braz. J. Microbiol. 2014, 45, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchakouteu, S.S.; Chatzifragkou, A.; Kalantzi, O.; Koutinas, A.A.; Aggelis, G.; Papanikolaou, S. Oleaginous yeast Cryptococcus curvatus exhibits interplay between biosynthesis of intracellular sugars and lipids. Eur. J. Lipid Sci. Technol. 2014, 117, 657–672. [Google Scholar] [CrossRef]
- Carota, E.; Crognale, S.; D’Annibale, A.; Gallo, A.M.; Stazi, S.R.; Petruccioli, M. A sustainable use of Ricotta Cheese Whey for microbial biodiesel production. Sci. Total Environ. 2017, 584–585, 554–560. [Google Scholar] [CrossRef]
- Miller, C.; Fosmer, A.; Rush, B.; McMullin, T.; Beacom, D.; Suominen, P. Industrial production of lactic acid. Compr. Biotechnol. 2011, 3, 179–188. [Google Scholar]
- Ryan, M.P.; Walsh, G. The biotechnological potential of whey. Rev. Environ. Sci. Bio/Technol. 2016, 15, 479–498. [Google Scholar] [CrossRef] [Green Version]
- Lane, M.M.; Morrissey, J.P. Kluyveromyces marxianus: A yeast emerging from its sister’s shadow. Fungal Biol. Rev. 2010, 24, 17–26. [Google Scholar] [CrossRef]
- Joshi, Y.; Senatore, B.; Poletto, M. Kluyveromyces marxianus biofilm in cheese whey fermentation for bioethanol production. Chem. Eng. Trans. 2011, 24, 493–498. [Google Scholar]
- Kachrimanidou, V.; Alimpoumpa, D.; Papadaki, A.; Lappa, I.; Alexopoulos, K.; Kopsahelis, N. Cheese whey utilization for biosurfactant production: Evaluation of bioprocessing strategies using novel Lactobacillus strains. Biom. Convers. Bioref. 2022, in press. [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part II: Technology and potential applications. Eur. J. Lipid Sci. Technol. 2011, 113, 1052–1073. [Google Scholar] [CrossRef]
- Bellou, S.; Triantaphyllidou, I.E.; Aggeli, D.; Elazzazy, A.M.; Baeshen, M.N.; Aggelis, G. Microbial oils as food additives: Recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 2016, 37, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Floetenmeyer, M.D.; Glatz, B.A.; Hammond, E.G. Continuous culture fermentation of whey permeate to produce microbial oil. J. Dairy Sci. 1985, 68, 633–637. [Google Scholar] [CrossRef]
- Ykema, A.; Verbree, E.C.; Kater, M.M.; Smit, H. Optimization of lipid production in the oleaginous yeast Apiotrichum curvatum in whey permeate. Appl. Microbiol. Biotechnol. 1988, 29, 211–218. [Google Scholar]
- Kopsahelis, N.; Dimou, C.; Papadaki, A.; Xenopoulos, E.; Kyraleou, M.; Kallithraka, S.; Kotseridis, Y.; Papanikolaou, S.; Koutinas, A.A. Refining of wine lees and cheese whey for the production of microbial oil, polyphenol-rich extracts and value-added co-products. J. Chem. Technol. Biotechnol. 2017, 93, 257–268. [Google Scholar] [CrossRef]
- Seo, Y.H.; Lee, I.; Jeon, S.H.; Han, J.I. Efficient conversion from cheese whey to lipid using Cryptococcus curvatus. Biochem. Eng. J. 2014, 90, 149–153. [Google Scholar] [CrossRef]
- Taskin, M.; Saghafian, A.; Aydogan, M.N.; Arslan, N.P. Microbial lipid production by cold-adapted oleaginous yeast Yarrowia lipolytica B9 in non-sterile whey medium. Biofuel. Bioprod. Bioref. 2015, 9, 595–605. [Google Scholar] [CrossRef]
- Arous, F.; Frikha, F.; Triantaphyllidou, I.-E.; Aggelis, G.; Nasri, M.; Mechichi, T. Potential utilization of agro-industrial wastewaters for lipid production by the oleaginous yeast Debaryomyces etchellsii. J. Clean. Prod. 2016, 133, 899–909. [Google Scholar] [CrossRef]
- Arous, F.; Atitallah, I.B.; Nasri, M.; Mechichi, T. A sustainable use of low-cost raw substrates for biodiesel production by the oleaginous yeast Wickerhamomyces anomalus. 3 Biotech 2017, 7, 268. [Google Scholar] [CrossRef]
- Vyas, S.; Chhabra, M. Assessing oil accumulation in the oleaginous yeast Cystobasidium oligophagum JRC1 using dairy waste cheese whey as a substrate. 3 Biotech 2019, 9, 173. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Chevalot, I.; Komaitis, M.; Aggelis, G.; Marc, I. Kinetic profile of the cellular lipid composition in an oleaginous Yarrowia lipolytica capable of producing a cocoa-butter substitute from industrial fats. Antonie Van Leeuwenhoek 2001, 80, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sarkany, N.; Cui, Y.; Blackburn, J.W. Batch stage study of lipid production from crude glycerol derived from yellow grease or animal fats through microagal fermentation. Bioresour. Technol. 2010, 101, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, D.; Psallida, C.; Sitarenious, P.; Flemetakis, E.; Diamantopoulou, P. Biochemical evaluation of Agaricus and Pleurotus strains in batch cultures for production optimization of valuable metabolites. Microorganisms 2022, 10, 964. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Sarantou, S.; Stoforos, N.G.; Kalantzi, O.; Papanikolaou, S. Biotechnological valorization of biodiesel-derived glycerol: Trials with the non-conventional yeasts Yarrowia lipolytica and Rhodosporidium sp. Carbon Resour. Convers. 2021, 4, 61–75. [Google Scholar] [CrossRef]
- Gardeli, C.; Athenaki, M.; Xenopoulos, E.; Mallouchos, A.; Koutinas, A.A.; Aggelis, G.; Papanikolaou, S. Lipid production and characterization by Mortierella (Umbelopsis) isabellina cultivated on lignocellulosic sugars. J. Appl. Microbiol. 2017, 123, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Lie, S. The EBC-ninhydrin method for determination of free alpha amino nitrogen. J. Inst. Brew. 1973, 79, 37–41. [Google Scholar] [CrossRef]
- ISO 8968-1. Milk–Determination of Nitrogen Content–Part 1: Kjeldahl Method. First Edition IDF 20-1. Available online: https://www.iso.org/standard/35120.html (accessed on 15 December 2001).
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Elnenaey, S.E.; Soliman, R. A sensitive colorimetric method for estimation of ascorbic acid. Talanta 1979, 26, 1164–1166. [Google Scholar] [CrossRef]
- Moreton, R.S. Physiology of lipid accumulating yeasts. In Single Cell Oil; Moreton, R.S., Ed.; Longman Scientific & Technical: Harlow, UK, 1988; pp. 1–32. [Google Scholar]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar]
- Papanikolaou, S.; Fakas, S.; Fick, M.; Chevalot, I.; Galiotou-Panayotou, M.; Komaitis, M.; Marc, I.; Aggelis, G. Biotechnological valorisation of raw glycerol discharged after bio-diesel (fatty acid methyl esters) manufacturing process: Production of 1,3-propanediol, citric acid and single cell oil. Biomass Bioenerg. 2008, 32, 60–71. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Sources of microbial oils with emphasis to Mortierella (Umbelopsis) isabellina fungus. World J. Microbiol. Biotechnol. 2019, 35, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Fakas, S.; Papanikolaou, S.; Galiotou-Panayotou, M.; Komaitis, M.; Aggelis, G. Lipids of Cunninghamella echinulata with emphasis to γ-linolenic acid distribution among lipid classes. Appl. Microbiol. Biotechnol. 2006, 73, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, A.; Kopsahelis, N.; Mallouchos, A.; Mandala, I.; Koutinas, A.A. Bioprocess development for the production of novel oleogels from soybean and microbial oils. Food Res. Int. 2019, 126, 108684. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Rontou, M.; Belka, A.; Athenaki, M.; Gardeli, C.; Mallouchos, A.; Kalantzi, O.; Koutinas, A.A.; Kookos, I.K.; Zeng, A.P.; et al. Conversion of biodiesel-derived glycerol into biotechnological products of industrial significance by yeast and fungal strains. Eng. Life Sci. 2017, 17, 262–281. [Google Scholar] [CrossRef]





| Total Solids (%) | Lactose (g/L) | FAN (mg/L) | Protein (Bradford) (g/L) | IP (g/L) | TKN (g/L) | pH |
|---|---|---|---|---|---|---|
| 7.2 ± 1.0 | 60.9 ± 1.8 | 63.0 ± 2.5 | 1.10 ± 0.15 | 0.68 ± 0.10 | 1.14 ± 0.13 | 6.25 ± 0.21 |
| Strain | Time (h) | Scons (g/L) | X (g/L) | L (g/L) | YX/S (g/g) | YL/S (g/g) | KL/X (g/g) | PL (g/L/h) |
|---|---|---|---|---|---|---|---|---|
| C. curvatus ATCC 20509 | 69 | 56.4 ± 3.1 | 22.0 ± 1.8 | 3.7 ± 0.4 | 0.39 | 0.07 | 0.17 | 0.054 |
| C. curvatus NRRL Y-1511 | 130 | 55.9 ± 2.0 | 15.5 ± 1.5 | 0.8 ± 0.2 | 0.28 | 0.01 | 0.05 | 0.006 |
| C. uzbekistanensis NRRL Y-44 | 115 | Tr. | n.g. | - | - | - | - | - |
| T. ovoides ACA-DC 5052 | 115 | Tr. | n.g. | - | -. | - | - | - |
| D. hansenii ACA-DC 5079 | 115 | 27.8 ± 1.4 | 5.1 ± 0.8 | 0.6 ± 0.2 | 0.19 | 0.02 | 0.12 | 0.005 |
| P. laurentii NRRL Y-2536 | 122 | 59.0 ± 2.0 | 16.5 ± 1.4 * | 2.0 ± 0.3 | 0.30 | 0.04 | 0.12 | 0.017 |
| P. laurentii NRRL YΒ-3594 | 115 | 54.7 ± 2.3 | 12.9 ± 1.8 | 1.2 ± 0.2 | 0.24 | 0.02 | 0.09 | 0.010 |
| C/N Moles/Moles | S0 (g/L) | Time (h) | Scons (g/L) | X (g/L) | L (g/L) | YX/S (g/g) | YL/S (g/g) | KL/X (g/g) | PL (g/L/h) |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 46.3 | 139 | 46.3 ± 2.0 | 20.0 ± 1.9 | 1.5 ± 0.2 | 0.43 | 0.03 | 0.08 | 0.011 |
| 124 | 58.3 | 96 | 58.0 ± 2.7 | 22.4 ± 2.2 | 7.3 ± 1.2 | 0.37 | 0.13 | 0.33 | 0.080 |
| 124 | 118.4 | 144 | 118.0 ± 3.6 | 44.0 ± 2.9 | 15.8 ± 1.6 | 0.38 | 0.13 | 0.36 | 0.110 |
| 337 | 107.0 | 111 | 36.0 ± 1.8 | 10.2 ± 1.8 | 4.2 ± 0.9 | 0.28 | 0.12 | 0.41 | 0.038 |
| 482 | 152.8 | 160 | 33.0 ± 2.1 | 14.9 ± 2.1 | 8.7 ± 1.6 | 0.45 | 0.26 | 0.58 | 0.054 |
| Strain | Time (h) | Scons (g/L) | X (g/L) | L (g/L) | YX/S (g/g) | YL/S (g/g) | KL/X (g/g) | PL (g/L/h) | FANcons (mg/L) | Proteincons (g/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| C. curvatus ATCC 20509 | 74 | 59.0 ± 0.8 | 22.4 ± 1.2 | 3.4 ± 0.2 | 0.38 | 0.06 | 0.15 | 0.045 | 31.6 ± 3.3 | 0.29 ± 0.08 |
| C. curvatus NRRL Y-1511 | 218 | 59.0 ± 2.0 | 20.6 ± 1.1 | 3.2 ± 0.1 | 0.35 | 0.05 | 0.16 | 0.015 | 36.0 ± 2.8 | 0.47 ± 0.10 |
| D. hansenii ACA-DC 5079 | 170 | 18.1 ± 1.9 | 8.3 ± 0.6 | 1.2 ± 0.2 | 0.40 | 0.02 | 0.15 | 0.007 | 41.1 ± 4.5 | 0.56 ± 0.08 |
| P. laurentii NRRL Y-2536 | 119 | 58.0 ± 2.2 | 22.0 ± 1.5 * | 1.2 ± 0.3 | 0.38 | 0.02 | 0.06 | 0.010 | 32.1 ± 4.0 | 0.13 ± 0.05 |
| P. laurentii NRRL YΒ-3594 | 101 | 55.7 ± 2.3 | 14.7 ± 1.4 | 1.0 ± 0.2 | 0.26 | 0.02 | 0.07 | 0.010 | 33.3 ± 2.9 | 0.09 ± 0.02 |
| Strain | g/100 g of Total FA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | Δ9C14:1 | C16:0 | Δ9C16:1 | C18:0 | Δ9C18:1 | Δ9,12C18:2 | SFA | UFA | MUFA | PUFA | |
| (a) | |||||||||||
| C. curvatus ATCC 20509 | 0.5 | 0.2 | 29.3 | 1.2 | 10.3 | 52.8 | 5.7 | 40.1 | 59.9 | 54.2 | 5.7 |
| C. curvatus NRRL Y-1511 | 0.8 | 1.2 | 26.3 | 0.2 | 14.9 | 47.3 | 9.3 | 42.0 | 58.0 | 48.7 | 9.3 |
| P. laurentii NRRL Y-2536 | 3.6 | 2.5 | 28.2 | 0.6 | 14.7 | 46.1 | 4.3 | 46.5 | 53.5 | 49.2 | 4.3 |
| P. laurentii NRRL YΒ-3594 | 0.9 | 0.5 | 19.1 | 0.3 | 14.4 | 56.4 | 8.3 | 34.5 | 65.5 | 57.2 | 8.3 |
| D. hansenii ACA-DC 5079 | 5.1 | 3.7 | 26.3 | 12.0 | 6.2 | 44.8 | 1.9 | 37.7 | 62.3 | 60.4 | 1.9 |
| (b) | |||||||||||
| C. curvatus ATCC 20509 | 0.7 | - | 36.5 | 1.0 | 10.4 | 46.6 | 4.8 | 47.6 | 52.4 | 47.6 | 4.8 |
| 0.5 | - | 24.3 | 0.6 | 14.3 | 53.7 | 5.7 | 39.9 | 60.1 | 54.4 | 5.7 | |
| Strain | Culture Conditions | C/N | Substrate | X (g/L) | L (g/L) | (g/g) | Biomass Productivity (g/L/h) | Lipids Productivity (g/L/h) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Apiotrichum curvatum ATCC 20509 | Batch b/r, pH = 4.8, t = 27 h | 25 | Ultrafiltrated CW Permeate C/N adjustment: NH4Cl | 23.20 | 4.2 | 0.18 | 0.86 | 0.155 | [36] |
| Batch b/r, pH = 4.8, t = 39 h | 40 | 21.60 | 5.6 | 0.36 | 0.56 | 0.199 | |||
| Batch b/r, pH = 4.8, t = 93 h | 70 | 19.70 | 11.4 | 0.58 | 0.21 | 0.123 | |||
| Fed-batch b/r, pH = 4.8, t = 70 h | 40 | 85.00 | 29.8 | 0.35 | 0.43 | 0.372 | |||
| Continuous b/r, pH = 4.8, D = 0.07 h−1 | 20 | 21.00 | 4.2 | 0.20 | 1.40 | 0.294 | |||
| Continuous b/r, pH = 4.8, D = 0.053 h−1 | 40 | 20.00 | 7.2 | 0.36 | 1.10 | 0.382 | |||
| Partial recycling b/r, pH = 4.8, D = 0.033 h−1 | 40 | 91.40 | 30.2 | 0.33 | 3.00 | 0.995 | |||
| Cryptococcus curvatus KCTC 27583 | Batch shaken baffled flasks, pH = 5.5, t = 24 h | n.a. | Alkaline Hydrodynamic Cavitation in CW | 7.20 | 4.7 | 0.65 | 0.30 | 0.195 | [38] |
| Cryptococcus curvatus NRRL Y-1511 | Batch shaken flasks, pH = 5.5, t = 72 h | 55 | Ricotta Second CW C/N adjustment: (NH4)2SO4 | 10.77 | 6.8 | 0.63 | 0.15 | 0.094 | [26] |
| Cryptococcus curvatus NRRL Y-1511 | Batch shaken flasks, pH = 5.5, t = 216 h | n.a. | Concentrated and deproteinized CW | 38.50 | 1.4 | 0.04 | 0.19 | 0.006 | [25] |
| Cryptococcus curvatus ATCC 20509 | Batch shaken flasks, pH = 6.0, t = 216 h | n.a. | Ultrafiltrated CW Permeate + Wines lees hydrolysate | 33.60 | 6.7 | 0.31 | 0.16 | 0.030 | [37] |
| Fed-batch b/r, pH = 6, t = 100h | n.a. | 66.80 | 33.1 | 0.50 | 0.67 | 0.490 | |||
| Candida curvata D | Batch b/r, pH = 5.4, t = 80 h | n.a. | Ultrafiltrated Cheddar CW Permeate | 13.80 | 7.6 | 0.55 | 0.17 | 0.095 | [35] |
| Continuous b/r, pH = 5.4, D = 0.02 h−1 | n.a. | 14.20 | 7.2 | 0.51 | 0.28 | 0.144 | |||
| Cryptococcus curvatus ATCC 20509 | Batch shaken flasks, pH = 5.5, t = 74 h | 58 | Centrifugated and Filtrated Mizithra Second CW | 22.40 | 3.3 | 0.15 | 0.30 | 0.045 | Current Study |
| Fed-Batch shaken flasks, pH = 5.5, t = 321 h | 182 | Centrifugated and Filtrated Mizithra Second CW supplemented with condensed CW derived lactose | 38.10 | 21.7 | 0.57 | 0.12 | 0.070 | ||
| Cryptococcus curvatus NRRL Y-1511 | Batch shaken flasks, pH = 5.5, t = 218 h | 58 | Centrifugated and Filtrated Mizithra Second CW | 20.60 | 3.2 | 0.16 | 0.09 | 0.015 | Current Study |
| Cryptococcus laurentii UCD 68-201 | Batch shaken flasks, pH = 5.5, t = 72 h | 55 | Ricotta Second CW C/N adjustment: (NH4)2SO4 | 7.28 | 5.1 | 0.70 | 0.10 | 0.070 | [26] |
| Batch Bioreactor pH = 5.5, t = 60 h | 14.37 | 9.9 | 0.69 | 0.24 | 0.165 | ||||
| Cryptococcus laurentii 11 | Batch shaken flasks, pH = 5.5, t = 240 h | n.a. | Centrifuged CW | 4.57 | 1.3 | 0.28 | 0.02 | 0.005 | [24] |
| Batch shaken flasks, pH = 6.5, t = 360 h | n.a. | Centrifuged CW + molasses | 16.58 | 1.5 | 0.09 | 0.05 | 0.004 | ||
| Papiliotrema laurentii NRRL Y-2536 | Batch shaken flasks, pH = 5.5, t = 119 h | 58 | Centrifugated and Filtrated Mizithra Second CW | 22.00 | 1.2 | 0.06 | 0.19 | 0.010 | Current Study |
| Papiliotrema laurentii NRRL YΒ-3594 | Batch Shaken flasks, pH = 5.5, t = 100.5 h | 58 | Centrifugated and Filtrated Mizithra Second CW | 14.70 | 1.0 | 0.07 | 0.15 | 0.010 | |
| Yarrowia lipolytica B9 | Batch shaken flasks, pH = 6.0, t = 120 h | n.a. | Deproteinized Cheese Broth | 7.40 | 4.3 | 0.58 | 0.06 | 0.035 | [39] |
| Cystobasidium oligophagum JRC1 | Batch shaken flasks, pH = 6.5, t = 168 h | ~60 | Untreated CW | 20.98 | 4.6 | 0.22 | 0.13 | 0.030 | [42] |
| 93 | Centrifugated and Filtrated Second CW | 12.79 | 5.6 | 0.44 | 0.08 | 0.033 | |||
| Debaryomyces etchellsii BM1 | Batch shaken flasks, pH = 6.0, t = 120 h | n.a. | Deproteinized CW | 3.90 | 0.4 | 0.10 | 0.03 | 0.003 | [40] |
| Debaryomyces hansenii hansenii ACA-DC 5079 | Batch shaken flasks, pH = 5.5, t = 170 h | 58 | Centrifugated and Filtrated Mizithra Second CW | 8.30 | 1.2 | 0.15 | 0.05 | 0.007 | Current Study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilakis, G.; Karayannis, D.; Massouras, T.; Politis, I.; Papanikolaou, S. Biotechnological Conversions of Mizithra Second Cheese Whey by Wild-Type Non-Conventional Yeast Strains: Production of Yeast Cell Biomass, Single-Cell Oil and Polysaccharides. Appl. Sci. 2022, 12, 11471. https://doi.org/10.3390/app122211471
Vasilakis G, Karayannis D, Massouras T, Politis I, Papanikolaou S. Biotechnological Conversions of Mizithra Second Cheese Whey by Wild-Type Non-Conventional Yeast Strains: Production of Yeast Cell Biomass, Single-Cell Oil and Polysaccharides. Applied Sciences. 2022; 12(22):11471. https://doi.org/10.3390/app122211471
Chicago/Turabian StyleVasilakis, Gabriel, Dimitris Karayannis, Theofilos Massouras, Ioannis Politis, and Seraphim Papanikolaou. 2022. "Biotechnological Conversions of Mizithra Second Cheese Whey by Wild-Type Non-Conventional Yeast Strains: Production of Yeast Cell Biomass, Single-Cell Oil and Polysaccharides" Applied Sciences 12, no. 22: 11471. https://doi.org/10.3390/app122211471
APA StyleVasilakis, G., Karayannis, D., Massouras, T., Politis, I., & Papanikolaou, S. (2022). Biotechnological Conversions of Mizithra Second Cheese Whey by Wild-Type Non-Conventional Yeast Strains: Production of Yeast Cell Biomass, Single-Cell Oil and Polysaccharides. Applied Sciences, 12(22), 11471. https://doi.org/10.3390/app122211471