Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release
Abstract
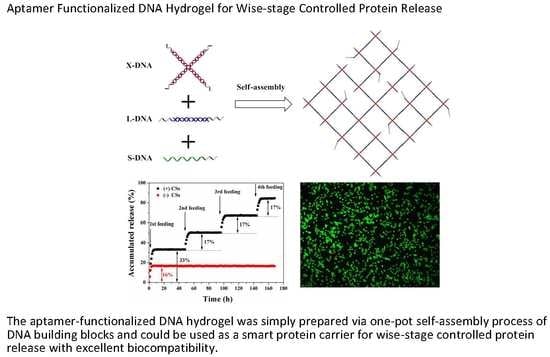
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Characterization of X and L-Shaped DNA Building Units
2.3. One-Pot Self-Assembly of Aptamer-Functionalized DNA Hydrogel
2.4. Determination of Mechanical Properties of the DNA Hydrogel
2.5. Aptamer-Functionalized DNA Hydrogel for Protein Capture
2.6. Aptamer-Functionalized DNA Hydrogel for Protein Release
2.7. Cell Culture
2.8. Statistical Analysis
3. Results and Discussion
3.1. The Preparation of DNA Building Units
3.2. Fabrication of Aptamer-Functionalized DNA Hydrogel
3.3. Examination of Thrombin Load by Aptamer-Functionalized DNA Hydrogel
3.4. Examination of Thrombin Release from Aptamer-Functionalized DNA Hydrogel
3.5. Application in Complex Matrixes
3.6. Cytocompatibility of the DNA Hydrogel
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sullivan, D.H.; Carter, W.J.; Warr, W.R.; Williams, L.H. Side effects resulting from the use of growth hormone and insulin-like growth factor-I as combined therapy to frail elderly patients. J. Gerontol. A Biol. Sci. Med. Sci. 1998, 7, 175–194. [Google Scholar] [CrossRef]
- Wang, D.; Hu, Y.; Liu, P.F.; Luo, D. Bioresponsive DNA hydrogels: Beyond the conventional stimuli responsiveness. Acc. Chem. Res. 2017, 50, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Lee, F.; Gao, S.J.; Chung, J.E.; Yano, H.; Kurisawa, M. Injectable hyaluronic acid-tyramine hydrogels incorporating interferon-α2a for liver cancer therapy. J. Control. Release 2013, 166, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Larson, D.R.; Lawrence, D.S. Illuminating the chemistry of life: Design, synthesis, and applications of “Caged” and related photoresponsive compounds. ACS Chem. Biol. 2009, 4, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, C.M.; Anseth, K.S. Hydrogels in healthcare: From static to dynamic material microenvironments. Acta Mater. 2013, 61, 931–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Metters, A.T. Metal-chelating affinity hydrogels for sustained protein release. J. Biomed. Mater. Res. A 2007, 83, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.R.; Wang, Y. Controlled delivery of heparin-binding EGF-like growth factor yields fast and comprehensive wound healing. J. Control. Release 2013, 166, 124–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.J.; McDonald, J.W., 3rd; Sakiyama-Elbert, S.E. Controlled release of neurotrophin-3 from fibrin gels for spinal cord injury. J. Control. Release 2004, 98, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Yang, W.; Kirn-Safran, C.B.; Farach-Carson, M.C.; Jia, X. Perlecan domain I-conjugated, hyaluronic acid-based hydrogel particles for enhanced chondrogenic differentiation via BMP-2 release. Biomaterials 2009, 30, 6964–6975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Liu, J.; Dvir, T.; Jin, L.; Tsui, J.H.; Qing, Q.; Suo, Z.; Langer, R.; Kohane, D.S.; Lieber, C.M. Macroporous nanowire nanoelectronic scaffolds for synthetic tissues. Nat. Mater. 2012, 11, 986–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, M.K.; Vermerris, W. Recent advances in nanomaterials for gene delivery-A review. Nanomaterials 2017, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Neff, D.; Green, N.; Norton, M.L. DNA origami reorganizes upon interaction with graphite: implications for high-resolution DNA directed protein patterning. Nanomaterials 2016, 6, 196. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Tapio, K.; Linko, V.; Kostiainen, M.A.; Toppari, J.J. Metallic nanostructures based on DNA nanoshapes. Nanomaterials 2016, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Finch, A.S.; Anton, C.M.; Jacob, C.M.; Proctor, T.J.; Stratis-Cullum, D.N. Assembly of DNA architectures in a non-aqueous solution. Nanomaterials 2012, 2, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Cao, T.Y.; Xu, Y.; Dong, Y.C.; Liu, D.S. Tuning the mechanical properties of a DNA hydrogel in three phases based on ATP aptamer. Int. J. Mol. Sci. 2018, 19, 1633. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jia, H.Y.; Cao, T.Y.; Liu, D.S. Supramolecular hydrogels based on DNA self-assembly. Acc. Chem. Res. 2017, 50, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Rinker, S.; Ke, Y.G.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nature Nanotechnol. 2008, 3, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Soontornworajit, B.; Zhou, J.; Shaw, M.T.; Fan, T.H.; Wang, Y. Hydrogel functionalization with DNA aptamers for sustained PDGF-BB release. Chem. Commun. 2010, 46, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Soontornworajit, B.; Zhou, J.; Snipes, M.P.; Battig, M.R.; Wang, Y. Affinity hydrogels for controlled protein release using nucleic acid aptamers and complementary oligonucleotides. Biomaterials 2011, 32, 6839–6849. [Google Scholar] [CrossRef] [PubMed]
- Battig, M.R.; Soontornworajit, B.; Wang, Y. Programmable release of multiple protein drugs from aptamer-functionalized hydrogels via nucleic acid hybridization. J. Am. Chem. Soc. 2012, 134, 12410–12413. [Google Scholar] [CrossRef] [PubMed]
- Soontornworajit, B.; Zhou, J.; Wang, Y. A hybrid particle–hydrogel composite for oligonucleotide-mediated pulsatile protein release. Soft Matter 2010, 6, 4255–4261. [Google Scholar] [CrossRef]
- Wang, Z.; Xia, J.; Cai, F.; Zhang, F.; Yang, M.; Bi, S.; Gui, R.; Li, Y.; Xia, Y. Aptamer-functionalized hydrogel as effective anti-cancer drugs delivery agents. Colloid. Surface B. 2015, 134, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Ohtsuki, S.; Araie, Y.; Umeki, Y.; Endo, M.; Emura, T.; Hidaka, K.; Sugiyama, H.; Takahashi, Y.; Takakura, Y.; et al. Self-assembling DNA hydrogel-based delivery of immunoinhibitory nucleic acids to immune cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Hu, Y.; Lu, C.-H.; Guo, W.; Aleman-Garcia, M.A.; Ricci, F.; Willner, I. pH-responsive and switchable triplex-based DNA hydrogels. Chem. Sci. 2015, 6, 4190–4195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, B.; Cheng, I.; Luo, K.Q.; Mi, Y. Capture and release of protein by a reversible DNA-induced sol-gel transition system. Angew. Chem. Int. Ed. Engl. 2008, 47, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhang, Z.; Soontornworajit, B.; Zhou, J.; Wang, Y. Cell adhesion on an artificial extracellular matrix using aptamer-functionalized PEG hydrogels. Biomaterials 2012, 33, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, N.; Zhang, Z.; Wang, Y. Endonuclease-responsive aptamer-functionalized hydrogel coating for sequential catch and release of cancer cells. Biomaterials 2013, 34, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Jiang, P.; Gaddes, E.R.; Zhao, N.; Abune, L.; Wang, Y. Aptamer-functionalized hydrogel for self-programmed protein release via sequential photoreaction and hybridization. Chem. Mater. 2017, 29, 5850–5857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Du, J.; Li, Y.; Wu, J.; Yu, F.; Chen, Y. An aptamer-patterned hydrogel for the controlled capture and release of proteins via biorthogonal click chemistry and DNA hybridization. J. Mater. Chem. B 2017, 5, 5974–5982. [Google Scholar] [CrossRef]
- Jin, C.; Zheng, J.; Li, C.; Qiu, L.; Zhang, X.; Tan, W. Aptamers selected by cell-SELEX for molecular imaging. J. Mol. Evol. 2015, 81, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; Lee, J.B.; Park, N.; Kwon, S.Y.; Umbach, C.C.; Luo, D. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, M.; Ogawa, K.; Umeki, Y.; Mohri, K.; Kawasaki, Y.; Watanabe, H.; Takahashi, N.; Kusuki, E.; Takahashi, R.; et al. Injectable, self-gelling, biodegradable, and immunomodulatory DNA hydrogel for antigen delivery. J. Control. Release 2014, 180, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.B.; Peng, S.; Yang, D.; Roh, Y.H.; Funabashi, H.; Park, N.; Rice, E.J.; Chen, L.; Long, R.; Wu, M.; et al. A mechanical metamaterial made from a DNA hydrogel. Nat. Nanotechnol. 2012, 7, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, Y.; Feng, F.; Shuang, S. A graphene oxide-based fluorescent aptasensor for alpha-fetoprotein detection. Anal. Methods 2016, 8, 6131–6134. [Google Scholar] [CrossRef]
- Cheng, E.; Xing, Y.; Chen, P.; Yang, Y.; Sun, Y.; Zhou, D.; Xu, L.; Fan, Q.; Liu, D. A pH-triggered, Fast-responding DNA hydrogel. Angew. Chem. Int. Ed. 2009, 48, 7796–7799. [Google Scholar] [CrossRef]
- Xing, Y.; Cheng, E.; Yang, Y.; Chen, P.; Zhang, T.; Sun, Y.; Yang, Z.; Liu, D. Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness. Adv. Mater. 2011, 23, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-J.; Kim, J.-T.; Kim, H.-J.; Kyung, H.-W.; Katila, P.; Lee, J.-H.; Yang, T.-H.; Yang, Y.-I.; Lee, S.-J. Epicardial delivery of VEGF and cardiac stem cells guided by 3-dimensional PLLA mat enhancing cardiac regeneration and angiogenesis in acute myocardial infarction. J. Control. Release 2015, 205, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, M.R.; Capelle, H.H.; Erber, R.; Ullrich, A.; Vajkoczy, P. Combined inhibition of vascular endothelial growth factor and platelet-derived growth factor signaling: Effects on the angiogenesis, microcirculation, and growth of orthotopic malignant gliomas. J. Neurosurg. 2005, 102, 363–370. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wei, B.; Mi, Y. Aptamer based reversible DNA induced hydrogel system for molecular recognition and separation. Chem. Commun. 2010, 46, 6308–6310. [Google Scholar] [CrossRef] [PubMed]
- Strehlitz, B.; Nikolaus, N.; Stoltenburg, R. Protein detection with aptamer biosensors. Sensors 2008, 8, 4296–4307. [Google Scholar] [CrossRef] [PubMed]
- Delport, F.; Pollet, J.; Janssen, K.; Verbruggen, B.; Knez, K.; Spasic, D.; Lammertyn, J. Real-time monitoring of DNA hybridization and melting processes using a fiber optic sensor. Nanotechnology 2012, 23, 065503. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xu, K.; Zheng, X.; Giacomin, A.J.; Mix, A.W.; Kao, W.J. 3D cell entrapment in crosslinked thiolated gelatin-poly (ethylene glycol) diacrylate hydrogels. Biomaterials 2012, 33, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, E.H.; Zanotelli, M.R.; Schwartz, M.P.; Murphy, W.L. Differential effects of cell adhesion, modulus and VEGFR-2 inhibition on capillary network formation in synthetic hydrogel arrays. Biomaterials 2014, 35, 2149–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Han, J.; Pei, Y.; Du, J. Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release. Appl. Sci. 2018, 8, 1941. https://doi.org/10.3390/app8101941
Liu C, Han J, Pei Y, Du J. Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release. Applied Sciences. 2018; 8(10):1941. https://doi.org/10.3390/app8101941
Chicago/Turabian StyleLiu, Chen, Jialun Han, Yuxuan Pei, and Jie Du. 2018. "Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release" Applied Sciences 8, no. 10: 1941. https://doi.org/10.3390/app8101941
APA StyleLiu, C., Han, J., Pei, Y., & Du, J. (2018). Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release. Applied Sciences, 8(10), 1941. https://doi.org/10.3390/app8101941