LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction †
Abstract
:Featured Application
Abstract
1. Introduction
- -
- LSTM python script enabling software verticalization and integration in ERP platforms oriented on patient management;
- -
- Integration of LSTM neural network into the information system collecting patient information and patient data;
- -
- Creation of different data models allowing data pre-processing and new facilities oriented on patient management;
- -
- Creation of a prediction model based on the simultaneous analysis of multiple attributes;
- -
- Adoption of artificial data in order to improve the training dataset;
- -
- Possibility to choose the best prediction models by reading different model outputs.
2. Materials and Methods
- PregnanciesNumber (PN): Pregnant number;
- GlucosePlasma (GP): Glucose concentration (after 2 h of oral glucose tolerance test);
- BloodPressureDiastolic (BPD): Blood pressure (mm Hg);
- SkinThicknessTriceps (STT): Skin fold thickness (mm);
- Insulin2-Hour (I): Serum insulin (mu U/mL);
- BMIBody (BMI): Mass index (weight in kg/(height in m)2);
- DiabetesPedigreeFunctionDiabetes (DPFD): Pedigree function;
- AgeAge (AA): Years old;
- OutcomeClass (OC): Binary variable (0 indicates the no-diabetes status of 268 samples, and 1 indicates the diabetes status of the remaining 500 cases of the training dataset).
3. Results
Training and Testing Dataset
- -
- Choose the attributes characterized by a higher correlation if compared with other attributes (in the case of study are insulin correlated with glucose, and skin thickness correlated with BMI);
- -
- Split the dataset for patients having diabetes or not (first partition);
- -
- The first partition has been furthermore split by considering the age (second partition);
- -
- The second partition is then split into a third one representing pregnant women (third partition);
- -
- Change of the correlated attributes by a low quantity of the values couple glucose and insulin (by increasing insulin is decreased the glucose of the same entity in order to balance the parameter variation), and skin thickness and BMI of the same person belonging to the same partition.
4. Discussion
- -
- Calculation of correlation matrix (analysis of correlation and weights between variables);
- -
- Check of 2D variable functions (check of samples dispersion);
- -
- Calculation of LSTM prediction of diabetic outcomes by changing the partitioning between the testing and the training dataset;
- -
- Choice procedures of the best LSTM model.
- -
- The extraction of outliers related to wrong measurements and to neglect from the training and testing dataset;
- -
- The combined analysis of the therapeutic plan of the monitored patient;
- -
- The analysis of possible failures of the adopted sensors;
- -
- A dynamical update of the training model by changing anomalous data records;
- -
- The digital traceability of the assistance pattern in order to choose a patient more suitable to construct the training model;
- -
- A pre-clustering of patients (data pre-processing performed by combining different attributes such as age, pathology, therapeutic plan, etc.).
- -
- Phase 1: Collecting patient data (by means of a well-structured database system allowing different data mining processing);
- -
- Phase 2: Pre-clustering and filtering of patient data (construction of a stable training dataset);
- -
- Phase 3: Pre-analysis of correlations between attributes and analysis of data dispersions;
- -
- Phase 4: Execution of the LSTM neural network algorithm by processing simultaneously different attributes (multi-attribute data processing);
- -
- Phase 5: Comparison of results by changing the testing dataset;
- -
- Phase 6: Choice of the best model to adopt following the analysis of phase 5.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
- # Visualize training history
- from keras.models import Sequential
- from keras.layers import LSTM
- from keras.layers import Dense
- import matplotlib.pyplot as plt
- import numpy
- from sklearn import preprocessing
- from sklearn.metrics import roc_curve
- from sklearn.metrics import roc_auc_score
- from matplotlib import pyplot
- # random seed (a random seed is fixed)
- seed = 42
- numpy.random.seed(seed)
- ‘dataset loading(csv format)’
- dataset = numpy.loadtxt(“C:/user /pime_indian_paper/dataset/diabetes3.csv”, delimiter = “,”)
- ‘Dataset Normalization’
- normalized = preprocessing.normalize(dataset, norm = ‘max’, axis = 0, copy = True)
- ‘Partitioning example: 80% as training set and the 20% of sample of the test dataset’
- X = normalized[:,0:8]
- Y = normalized[:,8]
- ‘Dataset structure: 1 column of eight row: time sequence data format’
- ‘We modify the dataset structure so that it has a column with 8 rows instead of a row with 8 columns (structure implementing a temporal sequence). For sequential dataset it is not considered the following reshaping’
- X = X.reshape(768, 8, 1)
- ‘LSTM model creation’
- ‘We will use an LSTM (Long Short Term Memory) network. Recurrent networks take as input not only the example of current input they see, but also what they have previously perceived. The decision taken by a recurrent network at the time t-1 influences the decision it will reach a moment later in time t: the recurrent networks have two sources of input, the present and the recent past. We will use on each neuron the RELU activation function that flattens the response to all negative values to zero, while leaving everything unchanged for values equal to or greater than zero (normalization)’
- model = Sequential()
- model.add(LSTM(32, input_shape = (8,1), return_sequences = True, kernel_initializer = ‘uniform’, activation = ‘relu’))
- model.add(LSTM(64, kernel_initializer = ‘uniform’, return_sequences = True, activation = ‘relu’ ))
- model.add(LSTM(128, kernel_initializer = ‘uniform’, activation = ‘relu’))
- model.add(Dense(256, activation = ‘relu’))
- model.add(Dense(128, activation = ‘relu’))
- model.add(Dense(64, activation = ‘relu’))
- model.add(Dense(16, activation = ‘relu’))
- model.add(Dense(1, activation = ‘sigmoid’))
- ‘Loss function’
- ‘We compile the model using as a NADAM optimizer that combines the peculiarities of the RMSProp optimizer with the momentum concept’
- ‘We calculate the loss function through the binary crossentropy’
- model.compile(loss = ‘binary_crossentropy’, optimizer = ‘NADAM’, metrics = [‘accuracy’])
- model.summary()
- # Fit the model
- history = model.fit(X, Y, validation_split = 0.33, epochs = 300, batch_size = 64, verbose = 1)
- ‘Graphical Reporting’
- plt.plot(history.history[‘acc’])
- plt.plot(history.history[‘val_acc’])
- plt.title(‘model accuracy’)
- plt.ylabel(‘accuracy’)
- plt.xlabel(‘epoch’)
- plt.legend([‘train’, ‘test’], loc = ‘upper left’)
- plt.savefig(‘accuracy.png’)
- plt.show()
- ‘Outputs plotting’
- plt.plot(history.history[‘loss’])
- plt.plot(history.history[‘val_loss’])
- plt.title(‘model loss’)
- plt.ylabel(‘loss’)
- plt.xlabel(‘epoch’)
- plt.legend([‘train’, ‘test’],loc = ‘upper left’)
- plt.savefig(‘loss.png’)
- plt.show()
- ‘model saving’
- model.save(‘pima_indian.model’)
- ‘Curva ROC Curve’
- probs = model.predict_proba(X)
- probs = probs[:,0]
- auc = roc_auc_score(Y, probs)
- print(‘AUC: %.3f’ % auc)
- fpr, tpr, thresholds = roc_curve(Y, probs)
- pyplot.plot([0, 1], [0, 1], linestyle = ‘--’)
- pyplot.plot(fpr, tpr, marker = ‘.’)
- pyplot.savefig(‘roc.png’)
- pyplot.show()
Appendix B

Appendix C
- model = Sequential()
- model.add(Dense(93, input_shape = (1200,), activation = ‘relu’))
- model.add(Dense(93, activation = ‘relu’))
- model.add(Dense(1, activation = ‘relu’))
- model.compile(metrics = [‘accuracy’, auroc], optimizer = Nadam(lr = 0.002, schedule_decay = 0.004), loss = ‘mean_squared_error’)
- model.summary()
| Layer (type) | Output Shape | Param # |
| dense_1 (Dense) | (None, 93) | 111693 |
| dense_2 (Dense) | (None, 93) | 8742 |
| dense_3 (Dense) | (None, 1) | 94 |
| Total params: 120,529 | ||
| Trainable params: 120,529 | ||
| Non-trainable params: 0 |
References
- Wimmer, H.; Powell, L.M. A comparison of open source tools for data science. J. Inf. Syst. Appl. Res. 2016, 9, 4–12. [Google Scholar]
- Al-Khoder, A.; Harmouch, H. Evaluating four of the most popular open source and free data mining tools. Int. J. Acad. Sci. Res. 2015, 3, 13–23. [Google Scholar]
- Gulli, A.; Pal, S. Deep Learning with Keras- Implement Neural Networks with Keras on Theano and TensorFlow; Birmingham- Mumbai Packt Book: Birmingham, UK, 2017; ISBN 978-1-78712-842-2. [Google Scholar]
- Kovalev, V.; Kalinovsky, A.; Kovalev, S. Deep learning with theano, torch, caffe, TensorFlow, and deeplearning4j: Which one is the best in speed and accuracy? In Proceedings of the XIII International Conference on Pattern Recognition and Information Processing, Minsk, Belarus, 3–5 October 2016; Belarus State University: Minsk, Belarus, 2016; pp. 99–103. [Google Scholar]
- Li, J.-S.; Yu, H.-Y.; Zhang, X.-G. Data mining in hospital information system. In New Fundamental Technologies in Data Mining; Funatsu, K., Ed.; Intech: London, UK, 2011. [Google Scholar]
- Goodwin, L.; VanDyne, M.; Lin, S. Data mining issues an opportunities for building nursing knowledge. J. Biomed. Inform. 2003, 36, 379–388. [Google Scholar] [CrossRef]
- Belacel, N.; Boulassel, M.R. Multicriteria fuzzy assignment method: A useful tool to assist medical diagnosis. Artif. Intell. Med. 2001, 21, 201–207. [Google Scholar] [CrossRef]
- Demšar, J.; Zupan, B.; Aoki, N.; Wall, M.J.; Granchi, T.H.; Beck, J.R. Feature mining and predictive model construction from severe trauma patient’s data. Int. J. Med. Inform. 2001, 36, 41–50. [Google Scholar] [CrossRef]
- Kusiak, A.; Dixon, B.; Shital, S. Predicting survival time for kidney dialysis patients: a data mining approach. Comput. Biol. Med. 2005, 35, 311–327. [Google Scholar] [CrossRef]
- Yu, H.-Y.; Li, J.-S. Data mining analysis of inpatient fees in hospital information system. In Proceedings of the IEEE International Symposium on IT in Medicine & Education (ITME2009), Jinan, China, 14–16 August 2009. [Google Scholar]
- Chae, Y.M.; Kim, H.S. Analysis of healthcare quality indicator using data mining and decision support system. Exp. Syst. Appl. 2003, 24, 167–172. [Google Scholar] [CrossRef]
- Morando, M.; Ponte, S.; Ferrara, E.; Dellepiane, S. Definition of motion and biophysical indicators for home-based rehabilitation through serious games. Information 2018, 9, 105. [Google Scholar] [CrossRef]
- Ozcan, Y.A. Quantitative Methods in Health Care Management, 2nd ed.; Josey-Bass: San Francisco, CA, USA, 2009; pp. 10–44. [Google Scholar]
- Ghavami, P.; Kapur, K. Artificial neural network-enabled prognostics for patient health management. In Proceedings of the IEEE Conference on Prognostics and Health Management (PHM), Denver, CO, USA, 18–21 June 2012. [Google Scholar]
- Grossi, E. Artificial neural networks and predictive medicine: A revolutionary paradigm shift. In Artificial Neural Networks—Methodological Advances and Biomedical Applications, 1st ed.; Suzuki, K., Ed.; InTech: Rijeka, Croatia, 2011; Volume 1, pp. 130–150. [Google Scholar]
- Adhikari, N.C.D. Prevention of heart problem using artificial intelligence. Int. J. Artif. Intell. Appl. 2018, 9, 21–35. [Google Scholar] [CrossRef]
- Galiano, A.; Massaro, A.; Boussahel, B.; Barbuzzi, D.; Tarulli, F.; Pellicani, L.; Renna, L.; Guarini, A.; De Tullio, G.; Nardelli, G.; et al. Improvements in haematology for home health assistance and monitoring by a web based communication system. In Proceedings of the IEEE International Symposium on Medical Measurements and Applications MeMeA, Benevento, Italy, 15–18 May 2016. [Google Scholar]
- Massaro, A.; Maritati, V.; Savino, N.; Galiano, A.; Convertini, D.; De Fonte, E.; Di Muro, M. A Study of a health resources management platform integrating neural networks and DSS telemedicine for homecare assistance. Information 2018, 9, 176. [Google Scholar] [CrossRef]
- Massaro, A.; Maritati, V.; Savino, N.; Galiano, A. Neural networks for automated smart health platforms oriented on heart predictive diagnostic big data systems. In Proceedings of the AEIT 2018 International Annual Conference, Bari, Italy, 3–5 October 2018. [Google Scholar]
- Saadatnejad, S.; Oveisi, M.; Hashemi, M. LSTM-based ECG classification for continuous monitoring on personal wearable devices. IEEE J. Biomed. Health Inform. 2019. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.; Tran, T.; Phung, D.; Venkatesh, S. Predicting healthcare trajectories from medical records: A deep learning approach. J. Biomed. Inform. 2017, 69, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Kaji, D.A.; Zech, J.R.; Kim, J.S.; Cho, S.K.; Dangayach, N.S.; Costa, A.B.; Oermann, E.K. An attention based deep learning model of clinical events in the intensive care unit. PLoS ONE 2019, 14, e0211057. [Google Scholar] [CrossRef] [PubMed]
- Pima Indians Diabetes Database. Available online: https://gist.github.com/ktisha/c21e73a1bd1700294ef790c56c8aec1f (accessed on 27 August 2019).
- Predict the Onset of Diabetes Based on Diagnostic Measures. Available online: https://www.kaggle.com/uciml/pima-indians-diabetes-database (accessed on 21 June 2019).
- Wu, H.; Yang, S.; Huang, Z.; He, J.; Wang, X. Type 2 diabetes mellitus prediction model based on data mining. Inform. Med. Unlocked 2018, 10, 100–107. [Google Scholar] [CrossRef]
- Luo, M.; Ke Wang, M.; Cai, Z.; Liu, A.; Li, Y.; Cheang, C.F. Using imbalanced triangle synthetic data for machine learning anomaly detection. Comput. Mater. Contin. 2019, 58, 15–26. [Google Scholar] [CrossRef]
- Al Helal, M.; Chowdhury, A.I.; Islam, A.; Ahmed, E.; Mahmud, S.; Hossain, S. An optimization approach to improve classification performance in cancer and diabetes prediction. In Proceedings of the International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’sBazar, Bangladesh, 7–9 February 2019. [Google Scholar]
- Li, T.; Fong, S. A fast feature selection method based on coefficient of variation for diabetics prediction using machine learning. Int. J. Extr. Autom. Connect. Health 2019, 1, 1–11. [Google Scholar] [CrossRef]
- Puneet, M.; Singh, Y.A. Impact of preprocessing methods on healthcare predictions. In Proceedings of the 2nd International Conference on Advanced Computing and Software Engineering (ICACSE), Sultanpur, India, 8–9 February 2019. [Google Scholar]
- Stranieri, A.; Yatsko, A.; Jelinek, H.F.; Venkatraman, S. Data-analytically derived flexi, le HbA1c thresholds for type 2 diabetes mellitus diagnostic. Artif. Intell. Res. 2016, 5, 111–134. [Google Scholar]
- Sudharsan, B.; Peeples, M.M.; Shomali, M.E. Hypoglycemia prediction using machine learning models for patients with type 2 diabetes. J. Diabetes Sci. Technol. 2015, 9, 86–90. [Google Scholar] [CrossRef]
- Mhaskar, H.N.; Pereverzyev, S.V.; Van Der Walt, M.D. A deep learning approach to diabetic blood glucose prediction. Front. Appl. Math. Stat. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Contreras, I.; Vehi, J. Artificial intelligence for diabetes management and decision support: Literature review. J. Med. Internet Res. 2018, 20, 1–24. [Google Scholar] [CrossRef]
- Bosnyak, Z.; Zhou, F.L.; Jimenez, J.; Berria, R. Predictive modeling of hypoglycemia risk with basal insulin use in type 2 diabetes: Use of machine learning in the LIGHTNING study. Diabetes Ther. 2019, 10, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Massaro, A.; Meuli, G.; Galiano, A. Intelligent electrical multi outlets controlled and activated by a data mining engine oriented to building electrical management. Int. J. Soft Comput. Artif. Intell. Appl. 2018, 7, 1–20. [Google Scholar] [CrossRef]
- Myers, J.L.; Well, A.D. Research Design and Statistical Analysis, 2nd ed.; Lawrence Erlbaum: Mahwah, NJ, USA, 2003. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.K.; Mihir, J.K.S.; Mohanty, N. Detection of diabetes using multilayer perceptron. In International Conference on Intelligent Computingand Applications, Advances in Intelligent Systems and Computing; Bhaskar, M.A., Dash, S.S., Das, S., Panigrahi, B.K., Eds.; Springer: Singapore, 2019. [Google Scholar]
- Swets, J.A. Measuring the accuracy of diagnostic systems. Science 1988, 240, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Diabetes Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Diabetes (accessed on 19 August 2019).
- Chui, K.T.; Fung, D.C.L.; Lytras, M.D. Predicting at-risk University students in a virtual learning environment via a machine learning algorithm. Comput. Hum. Behav. 2018, in press. [Google Scholar] [CrossRef]

















| PN | GP | BPD | STT | I | BMI | DPFD | AA | OC | |
|---|---|---|---|---|---|---|---|---|---|
| PN | 1 | 0.13 | 0.14 | −0.08 | −0.07 | 0.02 | −0.03 | 0.54 | 0.22 |
| GP | 0.13 | 1 | 0.15 | 0.06 | 0.03 | 0.22 | 0.14 | 0.26 | 0.47 |
| BPD | 0.14 | 0.15 | 1 | 0.21 | 0.09 | 0.28 | 0.04 | 0.24 | 0.07 |
| STT | −0.08 | 0.06 | 0.21 | 1 | 0.04 | 0.39 | 0.18 | −0.11 | 0.07 |
| I | −0.07 | 0.33 | 0.09 | 0.44 | 1 | 0.2 | 0.19 | −0.04 | 0.13 |
| BMI | 0.02 | 0.22 | 0.28 | 0.39 | 0.02 | 1 | 0.14 | 0.04 | 0.29 |
| DPFD | −0.03 | 0.14 | 0.04 | 0.18 | 0.19 | 0.14 | 1 | 0.03 | 0.17 |
| AA | 0.54 | 0.26 | 0.24 | −0.11 | −0.04 | 0.04 | 0.03 | 1 | 0.24 |
| OC | 0.22 | 0.47 | 0.07 | 0.07 | 0.1 | 0.29 | 0.17 | 0.24 | 1 |
| PN | GP | BPD | STT | I | BMI | DPFD | AA | OC (Predicted) |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | 0 |
| 3 | 78 | 50 | 32 | 88 | 31.0 | 0.248 | 26 | 1 |
| 10 | 115 | 0 | 0 | 0 | 35.3 | 0.134 | 29 | 0 |
| 2 | 197 | 70 | 45 | 543 | 30.5 | 0.158 | 53 | 1 |
| 8 | 125 | 96 | 0 | 0 | 0 | 0.232 | 54 | 1 |
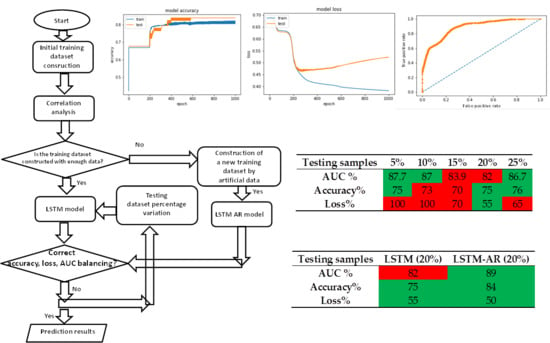
| Testing Samples | 5% | 10% | 15% | 20% | 25% |
|---|---|---|---|---|---|
| AUC % | 87.7 | 87 | 83.9 | 82 | 86.7 |
| Accuracy % | 75 | 73 | 70 | 75 | 76 |
| Loss % | 100 | 100 | 70 | 55 | 65 |
| Method | Test Set Accuracy % |
|---|---|
| MLP | 77.5 [38] |
| LSTM | 75 |
| LSTM-AR- | 84 |
| Testing Samples | LSTM (20%) | LSTM-AR (20%) |
|---|---|---|
| AUC % | 82 | 89 |
| Accuracy % | 75 | 84 |
| Loss % | 55 | 50 |
| AUC | AUC < 0.5 (50%) | AUC = 0.5 | 0.5 (50%) < AUC ≤ 0.7 (70%) | 0.7 (70%) < AUC ≤ 0.9 (90%) | 0.9 (90%) < AUC ≤ 1 (100%) | AUC = 1 |
|---|---|---|---|---|---|---|
| Classification of the discriminating capacity of a test | No sense test | Non-informative test | Inaccurate test | Moderately accurate test | Highly accurate test | Perfect test |
| Advantages | Limitations |
|---|---|
| DSS tool for diabetes prediction ready to use | Accurate training dataset |
| Multi attribute analysis | Redundancy of data processing (correlated attributes) |
| Reading procedure of outputs results | Presence of positive false and negative false due to wrong measurements |
| Choose of the best model according to simultaneous analyses (accuracy, loss, and AUC) | Finding a true compromise of efficiency parameter values |
| Network having a memory used for the data processing | It is necessary to acquire a correct temporal data sequence |
| Powerful approach if compared with ANN MLP method | High computational cost |
| Testing Samples | LSTM (20%) | LSTM-AR (20%) | MLP (20%) |
|---|---|---|---|
| AUC % | 91 | 91 | 94 |
| Accuracy % | 86 | 86 | 82 |
| Loss % | 10 | 10 | 14 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massaro, A.; Maritati, V.; Giannone, D.; Convertini, D.; Galiano, A. LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction. Appl. Sci. 2019, 9, 3532. https://doi.org/10.3390/app9173532
Massaro A, Maritati V, Giannone D, Convertini D, Galiano A. LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction. Applied Sciences. 2019; 9(17):3532. https://doi.org/10.3390/app9173532
Chicago/Turabian StyleMassaro, Alessandro, Vincenzo Maritati, Daniele Giannone, Daniele Convertini, and Angelo Galiano. 2019. "LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction" Applied Sciences 9, no. 17: 3532. https://doi.org/10.3390/app9173532
APA StyleMassaro, A., Maritati, V., Giannone, D., Convertini, D., & Galiano, A. (2019). LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction. Applied Sciences, 9(17), 3532. https://doi.org/10.3390/app9173532