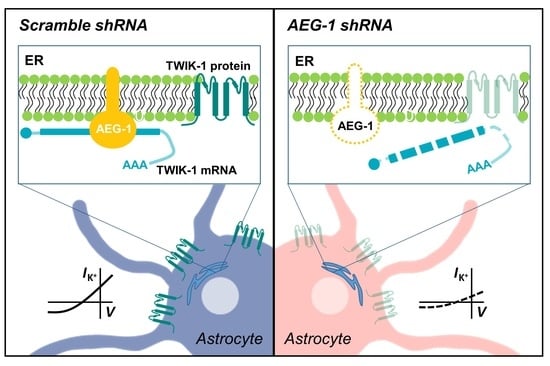
AEG-1 Regulates TWIK-1 Expression as an RNA-Binding Protein in Astrocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and shRNA
2.2. Primary Astrocyte Culture
2.3. RNA Sequencing (RNA-Seq) and Analysis
2.4. RT-PCR and qPCR
2.5. Immunocytochemistry
2.6. Electrophysiology
2.7. Western Blotting
2.8. RNA Immunoprecipitation (RIP) Assay
2.9. mRNA Stability Assays
2.10. Statistical Analysis
3. Results
3.1. AEG-1 Knockdown Down-Regulates TWIK-1 mRNA in Astrocytes
3.2. AEG-1 Knockdown Down-Regulates TWIK-1 Protein and TWIK-1-Mediated Potassium Currents in Cultured Astrocytes
3.3. AEG-1 Overexpression Up-Regulates TWIK-1 mRNA and Protein and Astrocytic Potassium Currents in Cultured Astrocytes
3.4. AEG-1 Is an RNA-Binding Protein That Enhances TWIK-1 mRNA Stability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kitchen, P.; Salman, M.M.; Halsey, A.M.; Clarke-Bland, C.; MacDonald, J.A.; Ishida, H.; Vogel, H.J.; Almutiri, S.; Logan, A.; Kreida, S.; et al. Targeting Aquaporin-4 Subcellular Localization to Treat Central Nervous System Edema. Cell 2020, 181, 784–799.e19. [Google Scholar] [CrossRef]
- Li, K.; Li, J.; Zheng, J.; Qin, S. Reactive Astrocytes in Neurodegenerative Diseases. Aging Dis. 2019, 10, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vartak-Sharma, N.; Ghorpade, A. Astrocyte elevated gene-1 regulates astrocyte responses to neural injury: Implications for reactive astrogliosis and neurodegeneration. J. Neuroinflamm. 2012, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.C.; Su, Z.Z.; Sarkar, D.; Emdad, L.; Volsky, D.J.; Fisher, P.B. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 2005, 353, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Emdad, L.; Sarkar, D.; Lee, S.G.; Su, Z.Z.; Yoo, B.K.; Dash, R.; Yacoub, A.; Fuller, C.E.; Shah, K.; Dent, P.; et al. Astrocyte elevated gene-1: A novel target for human glioma therapy. Mol. Cancer Ther. 2010, 9, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wu, J.; Ying, Z.; Chen, B.; Han, A.; Liang, Y.; Song, L.; Yuan, J.; Li, J.; Li, M. Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res. 2010, 70, 3750–3759. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.G.; Jeon, H.Y.; Su, Z.Z.; Richards, J.E.; Vozhilla, N.; Sarkar, D.; Van Maerken, T.; Fisher, P.B. Astrocyte elevated gene-1 contributes to the pathogenesis of neuroblastoma. Oncogene 2009, 28, 2476–2484. [Google Scholar] [CrossRef] [Green Version]
- Robertson, C.L.; Srivastava, J.; Rajasekaran, D.; Gredler, R.; Akiel, M.A.; Jariwala, N.; Siddiq, A.; Emdad, L.; Fisher, P.B.; Sarkar, D. The role of AEG-1 in the development of liver cancer. Hepat. Oncol. 2015, 2, 303–312. [Google Scholar] [CrossRef]
- Li, M.; Dai, Y.; Wang, L.; Li, L. Astrocyte elevated gene-1 promotes the proliferation and invasion of breast cancer cells by activating the Wnt/beta-catenin signaling pathway. Oncol. Lett. 2017, 13, 2385–2390. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhang, Z.; Wu, D.; Chen, H.; Qian, X.; Wang, X.; Huang, W. AEG-1 promotes the growth of gastric cancer through the upregulation of eIF4E expression. OncoTargets Ther. 2019, 12, 5887–5895. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Yang, Q.; Meng, F.; Shi, H.; Li, H.; Liang, Y.; Han, A. Astrocyte elevated gene-1 interacts with beta-catenin and increases migration and invasion of colorectal carcinoma. Mol. Carcinog. 2013, 52, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, X. The role of MTDH/AEG-1 in the progression of cancer. Int. J. Clin. Exp. Med. 2015, 8, 4795–4807. [Google Scholar] [PubMed]
- Yoo, B.K.; Emdad, L.; Lee, S.G.; Su, Z.Z.; Santhekadur, P.; Chen, D.; Gredler, R.; Fisher, P.B.; Sarkar, D. Astrocyte elevated gene-1 (AEG-1): A multifunctional regulator of normal and abnormal physiology. Pharmacol. Ther. 2011, 130, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emdad, L.; Sarkar, D.; Su, Z.Z.; Randolph, A.; Boukerche, H.; Valerie, K.; Fisher, P.B. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Res. 2006, 66, 1509–1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Guan, H.; Li, Y.; Ying, Z.; Wu, J.; Zhu, X.; Song, L.; Li, J.; Li, M. Astrocyte Elevated Gene 1 Interacts with Acetyltransferase p300 and c-Jun To Promote Tumor Aggressiveness. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.G.; Kim, K.; Kegelman, T.P.; Dash, R.; Das, S.K.; Choi, J.K.; Emdad, L.; Howlett, E.L.; Jeon, H.Y.; Su, Z.Z.; et al. Oncogene AEG-1 promotes glioma-induced neurodegeneration by increasing glutamate excitotoxicity. Cancer Res. 2011, 71, 6514–6523. [Google Scholar] [CrossRef] [Green Version]
- Hsu, J.C.C.; Reid, D.W.; Hoffman, A.M.; Sarkar, D.; Nicchitta, C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 2018, 24, 688–703. [Google Scholar] [CrossRef] [Green Version]
- Bethune, J.; Jansen, R.P.; Feldbrugge, M.; Zarnack, K. Membrane-Associated RNA-Binding Proteins Orchestrate Organelle-Coupled Translation. Trends Cell Biol. 2019, 29, 178–188. [Google Scholar] [CrossRef]
- Kwon, S.C.; Yi, H.; Eichelbaum, K.; Fohr, S.; Fischer, B.; You, K.T.; Castello, A.; Krijgsveld, J.; Hentze, M.W.; Kim, V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1122–1130. [Google Scholar] [CrossRef]
- Jagannathan, S.; Hsu, J.C.; Reid, D.W.; Chen, Q.; Thompson, W.J.; Moseley, A.M.; Nicchitta, C.V. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J. Biol. Chem. 2014, 289, 25907–25924. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, D.H.; Han, K.S.; Shim, J.W.; Yoon, B.E.; Kim, E.; Bae, J.Y.; Oh, S.J.; Hwang, E.M.; Marmorstein, A.D.; Bae, Y.C.; et al. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell 2012, 151, 25–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, E.M.; Kim, E.; Yarishkin, O.; Woo, D.H.; Han, K.S.; Park, N.; Bae, Y.; Woo, J.; Kim, D.; Park, M.; et al. A disulphide-linked heterodimer of TWIK-1 and TREK-1 mediates passive conductance in astrocytes. Nat. Commun. 2014, 5, 3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarishkin, O.; Lee, D.Y.; Kim, E.; Cho, C.H.; Choi, J.H.; Lee, C.J.; Hwang, E.M.; Park, J.Y. TWIK-1 contributes to the intrinsic excitability of dentate granule cells in mouse hippocampus. Mol. Brain 2014, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Yarishkin, O.; Kim, E.; Bae, Y.; Kim, A.; Kim, S.C.; Ryoo, K.; Cho, C.H.; Hwang, E.M.; Park, J.Y. TWIK-1/TASK-3 heterodimeric channels contribute to the neurotensin-mediated excitation of hippocampal dentate gyrus granule cells. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Moore, R.; Negishi, M. Nuclear receptor CAR specifically activates the two-pore K+ channel Kcnk1 gene in male mouse livers, which attenuates phenobarbital-induced hepatic hyperplasia. Toxicol. Sci. 2013, 132, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.; Jung, H.G.; Kim, S.C.; Choi, M.; Park, J.Y.; Lee, S.G.; Hwang, E.M. Astrocytic AEG-1 regulates expression of TREK-1 under acute hypoxia. Cell Biochem. Funct. 2020, 38, 167–175. [Google Scholar] [CrossRef]
- Lee, C.J.; Mannaioni, G.; Yuan, H.; Woo, D.H.; Gingrich, M.B.; Traynelis, S.F. Astrocytic control of synaptic NMDA receptors. J. Physiol. 2007, 581, 1057–1081. [Google Scholar] [CrossRef]
- Taylor, S.C.; Posch, A. The design of a quantitative western blot experiment. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Pillai-Kastoori, L.; Schutz-Geschwender, A.R.; Harford, J.A. A systematic approach to quantitative Western blot analysis. Anal. Biochem. 2020, 593. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Kelley, D.E. Inhibition of Rna Synthesis by Actinomycin-D—Characteristic Dose-Response of Different Rna Species. J. Cell. Physiol. 1970, 76, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [Green Version]
- Halbeisen, R.E.; Galgano, A.; Scherrer, T.; Gerber, A.P. Post-transcriptional gene regulation: From genome-wide studies to principles. Cell. Mol. Life Sci. 2008, 65, 798–813. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Ren, M.; Jiang, H.; Cui, S.; Wang, S.; Jiang, H.; Qi, Y.; Wang, J.; Wang, X.; Dong, G.; et al. Downregulated AEG-1 together with inhibited PI3K/Akt pathway is associated with reduced viability of motor neurons in an ALS model. Mol. Cell. Neurosci. 2015, 68, 303–313. [Google Scholar] [CrossRef]
- Leem, E.; Kim, H.J.; Choi, M.; Kim, S.; Oh, Y.S.; Lee, K.J.; Choe, Y.S.; Um, J.Y.; Shin, W.H.; Jeong, J.Y.; et al. Upregulation of neuronal astrocyte elevated gene-1 protects nigral dopaminergic neurons in vivo. Cell Death Dis. 2018, 9, 449. [Google Scholar] [CrossRef]
- Xia, C.; Fan, J.; Emanuel, G.; Hao, J.; Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA 2019, 116, 19490–19499. [Google Scholar] [CrossRef] [Green Version]




| Gene | Sequence |
|---|---|
| AEG-1 | |
| Forward Primer | 5′-CTATCTTCATCTACCCAGTTCCC-3′ |
| Probe | 5′-/56-FAM/CCTAGCTCA/ZEN/GACTGGAATGCACCA/31ABkFQ/-3′ |
| Reverse Primer | 5′-GGATGTTAGCCGTAATCAACCT-3′ |
| TWIK-1 | |
| Forward Primer | 5′-GCTACAACCAGAAGTTCCGA-3′ |
| Probe | 5′-/56-FAM/CCGAGGAGC/ZEN/AGGTAACACGTGAT/3IABkFQ/-3′ |
| Reverse Primer | 5′- TTCAGCTCGTGGAGTTCAC -3′ |
| GAPDH | |
| Forward Primer | 5′-GTGGAGTCATACTGGAACATGTAG-3′ |
| Probe | 5′-/56-FAM/TGCAAATGG/ZEN/CAGCCCTGGTG/3IABkFQ/-3′ |
| Reverse Primer | 5′-AATGGTGAAGGTCGGTGTG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.-G.; Kim, A.; Kim, S.-C.; Park, J.-Y.; Hwang, E.M. AEG-1 Regulates TWIK-1 Expression as an RNA-Binding Protein in Astrocytes. Brain Sci. 2021, 11, 85. https://doi.org/10.3390/brainsci11010085
Jung H-G, Kim A, Kim S-C, Park J-Y, Hwang EM. AEG-1 Regulates TWIK-1 Expression as an RNA-Binding Protein in Astrocytes. Brain Sciences. 2021; 11(1):85. https://doi.org/10.3390/brainsci11010085
Chicago/Turabian StyleJung, Hyun-Gug, Ajung Kim, Seung-Chan Kim, Jae-Yong Park, and Eun Mi Hwang. 2021. "AEG-1 Regulates TWIK-1 Expression as an RNA-Binding Protein in Astrocytes" Brain Sciences 11, no. 1: 85. https://doi.org/10.3390/brainsci11010085
APA StyleJung, H. -G., Kim, A., Kim, S. -C., Park, J. -Y., & Hwang, E. M. (2021). AEG-1 Regulates TWIK-1 Expression as an RNA-Binding Protein in Astrocytes. Brain Sciences, 11(1), 85. https://doi.org/10.3390/brainsci11010085