Effects of Serine or Threonine in the Active Site of Typical 2-Cys Prx on Hyperoxidation Susceptibility and on Chaperone Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Amplification, Cloning and Sequencing
2.2. Site Directed Mutagenesis
2.3. Oligonucleotides
2.4. Expression and Purification of Recombinant Proteins
2.5. NADPH Oxidation Assay
2.6. Western Blot Analysis to Evaluate the CP Hyperoxidation
2.7. In Silico Screening of the Typical 2-Cys Prx and Phylogenetic Analyses
2.8. Size Exclusion Chromatography (SEC) of Bacterial Typical 2-Cys Prx
2.9. Differential Scanning Fluorimetry
2.10. Chaperone Activity Assay
2.11. Analysis of Crystallographic Structures
3. Results
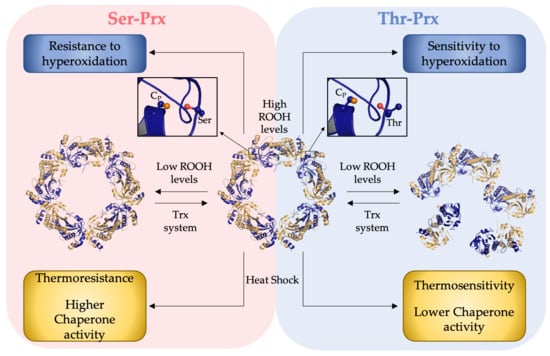
3.1. Presence of a Ser in the Catalytic Triad Increased Resistance of Yeast 2-Cys Prx to Hyperoxidation
3.2. Thr-Prx Are More Widely Distributed and Ser-Prx Is More Prevalent in Bacteria
3.3. Quaternary Structures Characterization of Bacterial Thr-Prx and Ser-Prx
3.4. Comparative Analysis of the Inactivation of Bacterial Thr-Prx and Ser-Prx by High Hydroperoxide Concentrations and by Thermal Insult
3.5. AhpC Containing Ser Presented Enhanced Chaperone Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Horta, B.B.; de Oliveira, M.A.; Discola, K.F.; Cussiol, J.R.; Netto, L.E. Structural and biochemical characterization of peroxiredoxin Qbeta from Xylella fastidiosa: Catalytic mechanism and high reactivity. J. Biol. Chem. 2010, 285, 16051–16065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogusucu, R.; Rettori, D.; Munhoz, D.C.; Netto, L.E.; Augusto, O. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: Rate constants by competitive kinetics. Free Radic. Biol. Med. 2007, 42, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Tairum, C.A.; Santos, M.C.; Breyer, C.A.; Geyer, R.R.; Nieves, C.J.; Portillo-Ledesma, S.; Ferrer-Sueta, G.; Toledo, J.C., Jr.; Toyama, M.H.; Augusto, O.; et al. Catalytic Thr or Ser Residue Modulates Structural Switches in 2-Cys Peroxiredoxin by Distinct Mechanisms. Sci. Rep. 2016, 6, 33133. [Google Scholar] [CrossRef] [Green Version]
- Truzzi, D.R.; Coelho, F.R.; Paviani, V.; Alves, S.V.; Netto, L.E.S.; Augusto, O. The bicarbonate/carbon dioxide pair increases hydrogen peroxide-mediated hyperoxidation of human peroxiredoxin 1. J. Biol. Chem. 2019, 294, 14055–14067. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Alm Rosenblad, M.; Molin, M.; Blomberg, A. Metagenomics reveals that detoxification systems are underrepresented in marine bacterial communities. BMC Genom. 2014, 15, 749. [Google Scholar] [CrossRef] [Green Version]
- Chae, H.Z.; Kim, H.J.; Kang, S.W.; Rhee, S.G. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res. Clin. Pract. 1999, 45, 101–112. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef]
- Wood, Z.A.; Schroder, E.; Robin Harris, J.; Poole, L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003, 28, 32–40. [Google Scholar] [CrossRef]
- Cox, A.G.; Winterbourn, C.C.; Hampton, M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2009, 425, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef]
- Brown, J.D.; Day, A.M.; Taylor, S.R.; Tomalin, L.E.; Morgan, B.A.; Veal, E.A. A peroxiredoxin promotes H2O2 signaling and oxidative stress resistance by oxidizing a thioredoxin family protein. Cell Rep. 2013, 5, 1425–1435. [Google Scholar] [CrossRef] [Green Version]
- Netto, L.E.; Antunes, F. The Roles of Peroxiredoxin and Thioredoxin in Hydrogen Peroxide Sensing and in Signal Transduction. Mol. Cells 2016, 39, 65–71. [Google Scholar]
- Rhee, S.G.; Woo, H.A. Multiple functions of peroxiredoxins: Peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid. Redox Signal. 2011, 15, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Stocker, S.; Maurer, M.; Ruppert, T.; Dick, T.P. A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol. 2018, 14, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.M.; Siu, K.L.; Wong, C.M.; Jin, D.Y. Loss of yeast peroxiredoxin Tsa1p induces genome instability through activation of the DNA damage checkpoint and elevation of dNTP levels. PLoS Genet. 2009, 5, e1000697. [Google Scholar] [CrossRef] [Green Version]
- Chae, H.Z.; Chung, S.J.; Rhee, S.G. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 1994, 269, 27670–27678. [Google Scholar] [CrossRef]
- Jonsson, T.J.; Ellis, H.R.; Poole, L.B. Cysteine reactivity and thiol-disulfide interchange pathways in AhpF and AhpC of the bacterial alkyl hydroperoxide reductase system. Biochemistry 2007, 46, 5709–5721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netto, L.E.S.; Chae, H.Z.; Kang, S.W.; Rhee, S.G.; Stadtman, E.R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J. Biol. Chem. 1996, 271, 15315–15321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pineyro, M.D.; Arcari, T.; Robello, C.; Radi, R.; Trujillo, M. Tryparedoxin peroxidases from Trypanosoma cruzi: High efficiency in the catalytic elimination of hydrogen peroxide and peroxynitrite. Arch. Biochem. Biophys. 2011, 507, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Parsonage, D.; Poole, L.B.; Karplus, P.A. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J. Mol. Biol. 2010, 402, 194–209. [Google Scholar] [CrossRef] [Green Version]
- Chae, H.Z.; Kim, I.H.; Kim, K.; Rhee, S.G. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J. Biol. Chem. 1993, 268, 16815–16821. [Google Scholar] [CrossRef]
- Flohe, L.; Budde, H.; Bruns, K.; Castro, H.; Clos, J.; Hofmann, B.; Kansal-Kalavar, S.; Krumme, D.; Menge, U.; Plank-Schumacher, K.; et al. Tryparedoxin peroxidase of Leishmania donovani: Molecular cloning, heterologous expression, specificity, and catalytic mechanism. Arch. Biochem. Biophys. 2002, 397, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Lotte, K.; Plessow, R.; Brockhinke, A.; Baier, M.; Dietz, K.J. Reaction mechanism of plant 2-Cys peroxiredoxin. Role of the C terminus and the quaternary structure. J. Biol. Chem. 2003, 278, 24409–24420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Vizuete, A.; Damdimopoulos, A.E.; Pedrajas, J.R.; Gustafsson, J.A.; Spyrou, G. Human mitochondrial thioredoxin reductase cDNA cloning, expression and genomic organization. Eur. J. Biochem. 1999, 261, 405–412. [Google Scholar] [CrossRef]
- Nagy, P.; Karton, A.; Betz, A.; Peskin, A.V.; Pace, P.; O’Reilly, R.J.; Hampton, M.B.; Radom, L.; Winterbourn, C.C. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: A kinetic and computational study. J. Biol. Chem. 2011, 286, 18048–18055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portillo-Ledesma, S.; Sardi, F.; Manta, B.; Tourn, M.V.; Clippe, A.; Knoops, B.; Alvarez, B.; Coitino, E.L.; Ferrer-Sueta, G. Deconstructing the catalytic efficiency of peroxiredoxin-5 peroxidatic cysteine. Biochemistry 2014, 53, 6113–6125. [Google Scholar] [CrossRef]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003, 425, 980–984. [Google Scholar] [CrossRef]
- Jonsson, T.J.; Johnson, L.C.; Lowther, W.T. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature 2008, 451, 98–101. [Google Scholar] [CrossRef]
- Day, A.M.; Brown, J.D.; Taylor, S.R.; Rand, J.D.; Morgan, B.A.; Veal, E.A. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell 2012, 45, 398–408. [Google Scholar] [CrossRef] [Green Version]
- Del Olmo, M.; Kramer, A.; Herzel, H. A Robust Model for Circadian Redox Oscillations. Int. J. Mol. Sci. 2019, 20, 2368. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veal, E.A.; Underwood, Z.E.; Tomalin, L.E.; Morgan, B.A.; Pillay, C.S. Hyperoxidation of Peroxiredoxins: Gain or Loss of Function? Antioxid. Redox Signal. 2018, 28, 574–590. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Kamariah, N.; Eisenhaber, B.; Eisenhaber, F.; Gruber, G. Molecular mechanism of the Escherichia coli AhpC in the function of a chaperone under heat-shock conditions. Sci. Rep. 2018, 8, 14151. [Google Scholar] [CrossRef]
- Moon, J.C.; Hah, Y.S.; Kim, W.Y.; Jung, B.G.; Jang, H.H.; Lee, J.R.; Kim, S.Y.; Lee, Y.M.; Jeon, M.G.; Kim, C.W.; et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J. Biol. Chem. 2005, 280, 28775–28784. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, F.; Tse, E.; Castro, H.; Makepeace, K.A.T.; Meinen, B.A.; Borchers, C.H.; Poole, L.B.; Bardwell, J.C.; Tomas, A.M.; Southworth, D.R.; et al. Chaperone activation and client binding of a 2-cysteine peroxiredoxin. Nat. Commun. 2019, 10, 659. [Google Scholar] [CrossRef]
- Hall, A.; Nelson, K.; Poole, L.B.; Karplus, P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011, 15, 795–815. [Google Scholar] [CrossRef] [Green Version]
- Peskin, A.V.; Dickerhof, N.; Poynton, R.A.; Paton, L.N.; Pace, P.E.; Hampton, M.B.; Winterbourn, C.C. Hyperoxidation of peroxiredoxins 2 and 3: Rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. J. Biol. Chem. 2013, 288, 14170–14177. [Google Scholar] [CrossRef] [Green Version]
- Wood, Z.A.; Poole, L.B.; Hantgan, R.R.; Karplus, P.A. Dimers to doughnuts: Redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry 2002, 41, 5493–5504. [Google Scholar] [CrossRef]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 2003, 300, 650–653. [Google Scholar] [CrossRef]
- Angelucci, F.; Saccoccia, F.; Ardini, M.; Boumis, G.; Brunori, M.; Di Leandro, L.; Ippoliti, R.; Miele, A.E.; Natoli, G.; Scotti, S.; et al. Switching between the alternative structures and functions of a 2-Cys peroxiredoxin, by site-directed mutagenesis. J. Mol. Biol. 2013, 425, 4556–4568. [Google Scholar] [CrossRef]
- Pastor-Flores, D.; Talwar, D.; Pedre, B.; Dick, T.P. Real-time monitoring of peroxiredoxin oligomerization dynamics in living cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16313–16323. [Google Scholar] [CrossRef]
- Dip, P.V.; Kamariah, N.; Nartey, W.; Beushausen, C.; Kostyuchenko, V.A.; Ng, T.S.; Lok, S.M.; Saw, W.G.; Eisenhaber, F.; Eisenhaber, B.; et al. Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim. Biophys. Acta 2014, 1837, 1932–1943. [Google Scholar] [CrossRef] [Green Version]
- Barranco-Medina, S.; Dietz, K.J. Thermodynamics of 2-Cys peroxiredoxin assembly determined by isothermal titration calorimetry. Methods Enzymol. 2009, 466, 409–430. [Google Scholar]
- Liebthal, M.; Kushwah, M.S.; Kukura, P.; Dietz, K.J. Single molecule mass photometry reveals dynamic oligomerization of plant and human peroxiredoxins for functional conservation and diversification. bioRxiv 2021. [Google Scholar] [CrossRef]
- Morais, M.A.; Giuseppe, P.O.; Souza, T.A.; Alegria, T.G.; Oliveira, M.A.; Netto, L.E.; Murakami, M.T. How pH modulates the dimer-decamer interconversion of 2-Cys peroxiredoxins from the Prx1 subfamily. J. Biol. Chem. 2015, 290, 8582–8590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, M.A.B.; Giuseppe, P.O.; Souza, T.; Castro, H.; Honorato, R.V.; Oliveira, P.S.L.; Netto, L.E.S.; Tomas, A.M.; Murakami, M.T. Calcium and magnesium ions modulate the oligomeric state and function of mitochondrial 2-Cys peroxiredoxins in Leishmania parasites. J. Biol. Chem. 2017, 292, 7023–7039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, A.; Balakrishna, A.M.; Nartey, W.; Kohlmeier, A.; Dip, P.V.; Bhushan, S.; Gruber, G. Atomic structure and enzymatic insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit C. Free Radic. Biol. Med. 2018, 115, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.J.; Perkins, A.; Van Swearingen, A.E.D.; Hartman, S.; Brereton, A.E.; Parsonage, D.; Salsbury, F.R., Jr.; Karplus, P.A.; Poole, L.B. Experimentally Dissecting the Origins of Peroxiredoxin Catalysis. Antioxid. Redox Signal. 2018, 28, 521–536. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, J.A.; Nelson, K.J.; Haynes, A.C.; Lee, J.; Reisz, J.A.; Graff, A.H.; Clodfelter, J.E.; Parsonage, D.; Poole, L.B.; Furdui, C.M.; et al. Novel hyperoxidation resistance motifs in 2-Cys peroxiredoxins. J. Biol. Chem. 2018, 293, 11901–11912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, M.A.; Discola, K.F.; Alves, S.V.; Medrano, F.J.; Guimaraes, B.G.; Netto, L.E. Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system. Biochemistry 2010, 49, 3317–3326. [Google Scholar] [CrossRef]
- Tairum, C.A., Jr.; de Oliveira, M.A.; Horta, B.B.; Zara, F.J.; Netto, L.E. Disulfide biochemistry in 2-cys peroxiredoxin: Requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin. J. Mol. Biol. 2012, 424, 28–41. [Google Scholar] [CrossRef]
- Gonzalez Porque, P.; Baldesten, A.; Reichard, P. Purification of a thioredoxin system from yeast. J. Biol. Chem. 1970, 245, 2363–2370. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, B.; Hecht, H.J.; Flohe, L. Peroxiredoxins. Biol. Chem. 2002, 383, 347–364. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Partch, C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015, 79, 21–28. [Google Scholar] [CrossRef]
- Kriznik, A.; Libiad, M.; Le Cordier, H.; Boukhenouna, S.; Toledano, M.B.; Rahuel-Clermont, S. Dynamics of a Key Conformational Transition in the Mechanism of Peroxiredoxin Sulfinylation. ACS Catal. 2020, 10, 3326–3339. [Google Scholar] [CrossRef]
- Munhoz, D.C.; Netto, L.E. Cytosolic thioredoxin peroxidase I and II are important defenses of yeast against organic hydroperoxide insult: Catalases and peroxiredoxins cooperate in the decomposition of H2O2 by yeast. J. Biol. Chem. 2004, 279, 35219–35227. [Google Scholar] [CrossRef] [Green Version]
- Zwick, J.V.; Noble, S.; Ellaicy, Y.K.; Coe, G.D.; Hakey, D.J.; King, A.N.; Sadauskas, A.J.; Faulkner, M.J. AhpA is a peroxidase expressed during biofilm formation in Bacillus subtilis. Microbiologyopen 2017, 6, e00403. [Google Scholar] [CrossRef] [Green Version]
- Peskin, A.V.; Meotti, F.C.; Kean, K.M.; Gobl, C.; Peixoto, A.S.; Pace, P.E.; Horne, C.R.; Heath, S.G.; Crowther, J.M.; Dobson, R.C.J.; et al. Modifying the resolving cysteine affects the structure and hydrogen peroxide reactivity of peroxiredoxin 2. J. Biol. Chem. 2021, in press. [Google Scholar] [CrossRef]
- Chuang, M.H.; Wu, M.S.; Lo, W.L.; Lin, J.T.; Wong, C.H.; Chiou, S.H. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. USA 2006, 103, 2552–2557. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.T.; Lee, S.S.; Mondal, S.; Tripathi, B.N.; Kim, S.; Lee, K.W.; Hong, S.H.; Bai, H.W.; Cho, J.Y.; Chung, B.Y. Enhancement of the Chaperone Activity of Alkyl Hydroperoxide Reductase C from Pseudomonas aeruginosa PAO1 Resulting from a Point-Specific Mutation Confers Heat Tolerance in Escherichia coli. Mol. Cells 2016, 39, 594–602. [Google Scholar] [CrossRef] [Green Version]
- Parsonage, D.; Nelson, K.J.; Ferrer-Sueta, G.; Alley, S.; Karplus, P.A.; Furdui, C.M.; Poole, L.B. Dissecting peroxiredoxin catalysis: Separating binding, peroxidation, and resolution for a bacterial AhpC. Biochemistry 2015, 54, 1567–1575. [Google Scholar] [CrossRef] [Green Version]
- Cha, M.K.; Bae, Y.J.; Kim, K.J.; Park, B.J.; Kim, I.H. Characterization of two alkyl hydroperoxide reductase C homologs alkyl hydroperoxide reductase C_H1 and alkyl hydroperoxide reductase C_H2 in Bacillus subtilis. World J. Biol. Chem. 2015, 6, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Charoenlap, N.; Eiamphungporn, W.; Chauvatcharin, N.; Utamapongchai, S.; Vattanaviboon, P.; Mongkolsuk, S. OxyR mediated compensatory expression between ahpC and katA and the significance of ahpC in protection from hydrogen peroxide in Xanthomonas campestris. FEMS Microbiol. Lett. 2005, 249, 73–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukumori, F.; Kishii, M. Molecular cloning and transcriptional analysis of the alkyl hydroperoxide reductase genes from Pseudomonas putida KT2442. J. Gen. Appl. Microbiol. 2001, 47, 269–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loprasert, S.; Atichartpongkun, S.; Whangsuk, W.; Mongkolsuk, S. Isolation and analysis of the Xanthomonas alkyl hydroperoxide reductase gene and the peroxide sensor regulator genes ahpC and ahpF-oxyR-orfX. J. Bacteriol. 1997, 179, 3944–3949. [Google Scholar] [CrossRef] [Green Version]
- Sherman, D.R.; Mdluli, K.; Hickey, M.J.; Arain, T.M.; Morris, S.L.; Barry, C.E., 3rd; Stover, C.K. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 1996, 272, 1641–1643. [Google Scholar] [CrossRef]
- Wilson, T.; de Lisle, G.W.; Marcinkeviciene, J.A.; Blanchard, J.S.; Collins, D.M. Antisense RNA to ahpC, an oxidative stress defence gene involved in isoniazid resistance, indicates that AhpC of Mycobacterium bovis has virulence properties. Microbiology 1998, 144, 2687–2695. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Gu, H.; Yang, Y.; Bai, H.; Zhao, C.; Si, M.; Su, T.; Shen, X. Molecular Mechanisms of AhpC in Resistance to Oxidative Stress in Burkholderia thailandensis. Front. Microbiol. 2019, 10, 1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Conover, R.C.; Benoit, S.; Olczak, A.A.; Olson, J.W.; Johnson, M.K.; Maier, R.J. Role of a bacterial organic hydroperoxide detoxification system in preventing catalase inactivation. J. Biol. Chem. 2004, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Species | Strain/Plasmid | Source |
|---|---|---|
| E. coli | ATCC25922 | ATCC * |
| E. coli | BL21 DE3/pET15b-ahpc (Pa) | This work |
| E. coli | BL21 DE3/pET15b-ahpc (Se) | This work |
| E. coli | BL21 DE3/pET15b-trx (Ec) | This work |
| E. coli | BL21 DE3/pET15b-trxr (Ec) | This work |
| E. coli | BL21 DE3/pET15b-tsa1 (Sc) | [52] |
| E. coli | BL21 DE3 Tuner/pET15b-tsa2 (Sc) | [3] |
| E. coli | BL21 DE3/pET15b-tsa1t44s (Sc) | [3] |
| E. coli | BL21 DE3 Tuner/pET15b-tsa2s44t (Sc) | This work |
| E. coli | BL21 DE3/pET15b-trx1 (Sc) | [51] |
| E. coli | BL21 DE3/pET15b-trxr1 (Sc) | [51] |
| E. coli | XL1-Blue | Agilent |
| P. aeruginosa | ATCC29853 | ATCC * |
| S. epidermidis | ATCC12228 | ATCC * |
| Oligonucleotide | Sequence |
|---|---|
| Ec_Trx_F | 5′ CGC GAT CCA TAT GAT GAG CGA TAA AAT TAT TCA CCT GAC T 3′ |
| Ec_Trx_R | 5′ CGC AAG CTT GGA TCC TTA CGC CAG GTT AGC GTC GAG GAA 3′ |
| Ec_TrxR_F | 5′ CGC GAT CCA TAT GAT GGG CAC GAC CAA ACA CAG TAA ACT G 3′ |
| Ec_TrxR_R | 5′ CGC AAG CTT GGA TCC TTA TTT TGC GTC AGC TAA ACC ATC 3′ |
| Pa_AhpC_ F | 5′ CGC GAT CCA TAT GAT GTC CCT GAT CAA CAC TCA AGT CCA A 3′ |
| Pa_AhpC_R | 5′ CGC AAG CTT GGA TCC TTA GAT CTT GCC GAC CAG GTC CAG G 3′ |
| Pa_AhpC_T43S_F | 5′ GCT GCC TTC TCC TTC AAC TGC 3′ |
| Pa_AhpC_T43S_R | 5′ GCA GTT GAA GGA GAA GGC AGC 3′ |
| Se_AhpC_S46T_F | 5′ GCG GAC TTC ACC TTT GTT TGC 3′ |
| Se_AhpC_S46T_R | 5′ GCA AAC AAA GGT GAA GTC CGC 3′ |
| Tsa2S44T_F | 5′ ATT GGC TTT TAC TTT TGT CTG TC 3′ |
| Tsa2S44T_R | 5′ GAC AGA CAA AAG TAA AAG CCA AT 3′ |
| Thr | Ser | ||||||
|---|---|---|---|---|---|---|---|
| Bacteria | Eukarya | Bacteria | Eukarya | ||||
| Acidobacteria | 15 | Alveolata | 107 | FCB group * | 14 | Amoebozoa | 1 |
| Aquificae | 16 | Amoebozoa | 83 | Proteobacteria | 15 | Euglenozoa | 12 |
| Calditrichaeota | 2 | Apusozoa | 2 | PVC group § | 12 | Opisthokonta | 75 |
| Chrysiogenetes | 1 | Cryptophyta | 1 | Spirochaetes | 2 | Rhodophyta | 2 |
| Deferribacteres | 5 | Euglenozoa | 105 | Synergistetes | 1 | ||
| Elusimicrobia | 18 | Heterolobosea | 1 | Terrabacteria group | 803 | ||
| Environmental samples | 3 | Opisthokonta | 2079 | Unclassified | 4 | ||
| FCB group * | 1455 | Parabasalia (parabasalids) | 13 | ||||
| Fusobacteria | 78 | Rhizaria | 4 | ||||
| Nitrospinae/Tectomicrobia group | 11 | Rhodophyta | 116 | ||||
| Nitrospirae | 15 | Stramenopiles | 136 | ||||
| Proteobacteria | 4890 | Viridiplantae | 302 | ||||
| PVC group | 220 | ||||||
| Spirochaetes | 149 | ||||||
| Synergistetes | 10 | ||||||
| Terrabacteria group | 3564 | ||||||
| Thermodesulfobacteria | 2 | ||||||
| Unclassified | 245 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tairum, C.A.; Santos, M.C.; Breyer, C.A.; de Oliveira, A.L.P.; Cabrera, V.I.M.; Toledo-Silva, G.; Mori, G.M.; Toyama, M.H.; Netto, L.E.S.; de Oliveira, M.A. Effects of Serine or Threonine in the Active Site of Typical 2-Cys Prx on Hyperoxidation Susceptibility and on Chaperone Activity. Antioxidants 2021, 10, 1032. https://doi.org/10.3390/antiox10071032
Tairum CA, Santos MC, Breyer CA, de Oliveira ALP, Cabrera VIM, Toledo-Silva G, Mori GM, Toyama MH, Netto LES, de Oliveira MA. Effects of Serine or Threonine in the Active Site of Typical 2-Cys Prx on Hyperoxidation Susceptibility and on Chaperone Activity. Antioxidants. 2021; 10(7):1032. https://doi.org/10.3390/antiox10071032
Chicago/Turabian StyleTairum, Carlos A., Melina Cardoso Santos, Carlos Alexandre Breyer, Ana Laura Pires de Oliveira, Vitoria Isabela Montanhero Cabrera, Guilherme Toledo-Silva, Gustavo Maruyama Mori, Marcos Hikari Toyama, Luis Eduardo Soares Netto, and Marcos Antonio de Oliveira. 2021. "Effects of Serine or Threonine in the Active Site of Typical 2-Cys Prx on Hyperoxidation Susceptibility and on Chaperone Activity" Antioxidants 10, no. 7: 1032. https://doi.org/10.3390/antiox10071032
APA StyleTairum, C. A., Santos, M. C., Breyer, C. A., de Oliveira, A. L. P., Cabrera, V. I. M., Toledo-Silva, G., Mori, G. M., Toyama, M. H., Netto, L. E. S., & de Oliveira, M. A. (2021). Effects of Serine or Threonine in the Active Site of Typical 2-Cys Prx on Hyperoxidation Susceptibility and on Chaperone Activity. Antioxidants, 10(7), 1032. https://doi.org/10.3390/antiox10071032