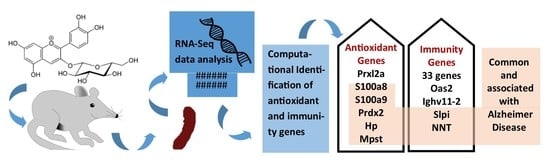
Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Animal Model
2.2. RNA-Seq Library Preparation and Sequencing
2.3. RNA-Seq Analysis of Assembly and Differential Expression of Genes
2.4. Function Enrichment of DEGs
2.5. Comparison of DEGs and Identification of Antioxidant and Immune Genes
2.6. Protein–Protein Interaction Network
2.7. Quantitative Real-Time PCR (qRT-PCR) Assay to Validate the Expression of Important Genes
3. Results
3.1. Quality Control and Alignment of RNA-Seq Reads
3.2. Assembly and Differential Expression Analysis
3.3. Functional Enrichment Analysis of DEGs
3.4. Identification of Common DEGs in Comparison
3.5. Identification of DEGs with Antioxidant Activity
3.6. Identification of DEGs with Immune-Related Function
3.7. PPIN Analysis
3.8. qRT-PCR Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jia, Y.; Wu, C.; Kim, Y.-S.; Yang, S.O.; Kim, Y.; Kim, J.-S.; Jeong, M.-Y.; Lee, J.H.; Kim, B.; Lee, S. A dietary anthocyanin cyanidin-3-O-glucoside binds to PPARs to regulate glucose metabolism and insulin sensitivity in mice. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Li, W.; Chen, S.; Zhou, G.; Li, H.; Zhong, L.; Liu, S. Potential role of cyanidin 3-glucoside (C3G) in diabetic cardiomyopathy in diabetic rats: An in vivo approach. Saudi J. Biol. Sci. 2018, 25, 500–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Zhai, Q.; Li, Y.; Cao, M.; Xu, Y.; Zhao, K.; Wang, T. Cyanidin-3-O-glucoside ameliorates diabetic nephropathy through regulation of glutathione pool. Biomed. Pharmacother. 2018, 103, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-M.; Lee, H.S.; Jung, J.I.; Kim, S.M.; Kim, N.Y.; Seo, T.S.; Bae, J.-S.; Kim, E.J. Cyanidin-3-O-galactoside-enriched Aronia melanocarpa extract attenuates weight gain and adipogenic pathways in high-fat diet-induced obese C57BL/6 mice. Nutrients 2019, 11, 1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloud, B.M.; Raj, P.; McCallum, J.; Kirby, C.; Louis, X.L.; Jahan, F.; Yu, L.; Hiebert, B.; Duhamel, T.A.; Wigle, J.T. Cyanidin 3-O-glucoside prevents the development of maladaptive cardiac hypertrophy and diastolic heart dysfunction in 20-week-old spontaneously hypertensive rats. Food Funct. 2018, 9, 3466–3480. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, F.; Wang, W.; Sang, J.; Jia, L.; Li, L.; Lu, F. Cyanidin-3-O-glucoside inhibits Aβ40 fibrillogenesis, disintegrates preformed fibrils, and reduces amyloid cytotoxicity. Food Funct. 2020, 11, 2573–2587. [Google Scholar] [CrossRef]
- Ma, B.; Wu, Y.; Chen, B.; Yao, Y.; Wang, Y.; Bai, H.; Li, C.; Yang, Y.; Chen, Y. Cyanidin-3-O-β-glucoside attenuates allergic airway inflammation by modulating the IL-4Rα-STAT6 signaling pathway in a murine asthma model. Int. Immunopharmacol. 2019, 69, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wang, X.; Liu, Y.; Xia, M. Supplementation with cyanidin-3-O-β-glucoside protects against hypercholesterolemia-mediated endothelial dysfunction and attenuates atherosclerosis in apolipoprotein E–deficient mice. J. Nutr. 2012, 142, 1033–1037. [Google Scholar] [CrossRef] [Green Version]
- Amararathna, M.; Hoskin, D.W.; Rupasinghe, H. Cyanidin-3-O-Glucoside-Rich Haskap Berry Administration Suppresses Carcinogen-Induced Lung Tumorigenesis in A/JCr Mice. Molecules 2020, 25, 3823. [Google Scholar] [CrossRef]
- Mazewski, C.; Kim, M.S.; de Mejia, E.G. Anthocyanins, delphinidin-3-O-glucoside and cyanidin-3-O-glucoside, inhibit immune checkpoints in human colorectal cancer cells in vitro and in silico. Sci. Rep. 2019, 9, 11560. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Du, Y.; Li, H.; Wang, L.; Ponikwicka-Tyszko, D.; Lebiedzinska, W.; Pilaszewicz-Puza, A.; Liu, H.; Zhou, L.; Fan, H. Cyanidin-3-o-glucoside pharmacologically inhibits tumorigenesis via estrogen receptor β in melanoma mice. Front. Oncol. 2019, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Jongsomchai, K.; Leardkamolkarn, V.; Mahatheeranont, S. A rice bran phytochemical, cyanidin 3-glucoside, inhibits the progression of PC3 prostate cancer cell. Anat. Cell Biol. 2020, 53, 481. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yuan, M.; Ye, Q.; Wang, X.; Xu, J.; Shi, G.; Hu, Z. Cyanidin-3-O-glucoside inhibits epithelial-to-mesenchymal transition, and migration and invasion of breast cancer cells by upregulating KLF4. Food Nutr. Res. 2020, 64. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L. Cyanidin-3-glucoside inhibits inflammatory activities in human fibroblast-like synoviocytes and in mice with collagen-induced arthritis. Clin. Exp. Pharmacol. Physiol. 2018, 45, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Sukprasansap, M.; Chanvorachote, P.; Tencomnao, T. Cyanidin-3-glucoside activates Nrf2-antioxidant response element and protects against glutamate-induced oxidative and endoplasmic reticulum stress in HT22 hippocampal neuronal cells. BMC Complementary Med. Ther. 2020, 20, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, D.; Cimino, F.; Fratantonio, D.; Molonia, M.S.; Bashllari, R.; Busà, R.; Saija, A.; Speciale, A. Cyanidin-3-O-glucoside modulates the in vitro inflammatory crosstalk between intestinal epithelial and endothelial cells. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef]
- Shin, H.-Y.; Kim, J.; Lee, S.; Park, M.S.; Park, S.; Huh, S. Cause-of-death statistics in 2018 in the Republic of Korea. J. Korean Med Assoc./Taehan Uisa Hyophoe Chi 2020, 63, 286–297. [Google Scholar]
- 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [CrossRef]
- 2021 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2021, 17, 327–406. [CrossRef]
- Yang, J.S.; Perveen, S.; Ha, T.J.; Kim, S.Y.; Yoon, S.H. Cyanidin-3-glucoside inhibits glutamate-induced Zn2+ signaling and neuronal cell death in cultured rat hippocampal neurons by inhibiting Ca2+-induced mitochondrial depolarization and formation of reactive oxygen species. Brain Res. 2015, 1606, 9–20. [Google Scholar] [CrossRef]
- Bhuiyan, M.I.H.; Kim, H.-B.; Kim, S.Y.; Cho, K.-O. The neuroprotective potential of cyanidin-3-glucoside fraction extracted from mulberry following oxygen-glucose deprivation. Korean J. Physiol. Pharmacol. Off. J. Korean Physiol. Soc. Korean Soc. Pharmacol. 2011, 15, 353. [Google Scholar] [CrossRef] [Green Version]
- Kang, T.H.; Hur, J.Y.; Kim, H.B.; Ryu, J.H.; Kim, S.Y. Neuroprotective effects of the cyanidin-3-O-β-d-glucopyranoside isolated from mulberry fruit against cerebral ischemia. Neurosci. Lett. 2006, 391, 122–126. [Google Scholar] [CrossRef]
- Ke, Z.; Liu, Y.; Wang, X.; Fan, Z.; Chen, G.; Xu, M.; Bower, K.A.; Frank, J.A.; Ou, X.; Shi, X. Cyanidin-3-glucoside ameliorates ethanol neurotoxicity in the developing brain. J. Neurosci. Res. 2011, 89, 1676–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.S.; Jeon, S.; Yoon, K.D.; Yoon, S.H. Cyanidin-3-glucoside inhibits amyloid β25–35-induced neuronal cell death in cultured rat hippocampal neurons. Korean J. Physiol. Pharmacol. Off. J. Korean Physiol. Soc. Korean Soc. Pharmacol. 2018, 22, 689. [Google Scholar] [CrossRef] [Green Version]
- Tarozzi, A.; Merlicco, A.; Morroni, F.; Franco, F.; Cantelli-Forti, G.; Teti, G.; Falconi, M.; Hrelia, P. Cyanidin 3-O-glucopyranoside protects and rescues SH-SY5Y cells against amyloid-beta peptide-induced toxicity. Neuroreport 2008, 19, 1483–1486. [Google Scholar] [CrossRef] [PubMed]
- Tarozzi, A.; Morroni, F.; Merlicco, A.; Bolondi, C.; Teti, G.; Falconi, M.; Cantelli-Forti, G.; Hrelia, P. Neuroprotective effects of cyanidin 3-O-glucopyranoside on amyloid beta (25–35) oligomer-induced toxicity. Neurosci. Lett. 2010, 473, 72–76. [Google Scholar] [CrossRef]
- Song, N.; Zhang, L.; Chen, W.; Zhu, H.; Deng, W.; Han, Y.; Guo, J.; Qin, C. Cyanidin 3-O-β-glucopyranoside activates peroxisome proliferator-activated receptor-γ and alleviates cognitive impairment in the APPswe/PS1ΔE9 mouse model. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 1786–1800. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhang, J.; Qin, M. Protective effect of cyanidin 3-O-glucoside on beta-amyloid peptide-induced cognitive impairment in rats. Neurosci. Lett. 2013, 534, 285–288. [Google Scholar] [PubMed]
- Frost, G.R.; Jonas, L.A.; Li, Y.-M. Friend, foe or both? Immune activity in Alzheimer’s disease. Front. Aging Neurosci. 2019, 11, 337. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019, 4, eaau6085. [Google Scholar] [CrossRef]
- Gu, X.; Ma, Z.; Fang, J.; Cai, D.; Zuo, Z.; Liang, S.; Cui, H.; Deng, J.; Ma, X.; Ren, Z. Obesity enhances antioxidant capacity and reduces cytokine levels of the spleen in mice to resist splenic injury challenged by Escherichia coli. J. Immunol. Res. 2020, 2020, 5948256-13. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Li, Y.; An, J.; Liu, Z.; Chen, Q. Antioxidative and inflammatory responses in spleen and head kidney of yellow catfish (pelteobagrus fulvidraco) induced by waterborne cadmium exposure. Turk. J. Fish. Aquat. Sci. 2019, 20, 87–96. [Google Scholar]
- Zhang, M.; Luo, J.; Zhang, C.; Cao, H.; Xia, B.; Hu, G. Alterations in antioxidant function and cell apoptosis in duck spleen exposed to molybdenum and/or cadmium. J. Vet. Sci. 2017, 18, 193. [Google Scholar] [CrossRef]
- Omar, H.E.-D.M.; Eldien, H.M.S.; Badary, M.S.; Al-Khatib, B.Y.; AbdElgaffar, S.K. The immunomodulating and antioxidant activity of fucoidan on the splenic tissue of rats treated with cyclosporine A. J. Basic Appl. Zool. 2013, 66, 243–254. [Google Scholar] [CrossRef] [Green Version]
- Chawla, A.; Stobdan, T.; Srivastava, R.B.; Jaiswal, V.; Chauhan, R.S.; Kant, A. Sex-biased temporal gene expression in male and female floral buds of seabuckthorn (Hippophae rhamnoides). PLoS ONE 2015, 10, e0124890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guleria, V.; Jaiswal, V. Comparative transcriptome analysis of different stages of Plasmodium falciparum to explore vaccine and drug candidates. Genomics 2020, 112, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Chauhan, R.S.; Rout, C. Common antigens prediction in bacterial bioweapons: A perspective for vaccine design. Infect. Genet. Evol. 2014, 21, 315–319. [Google Scholar] [CrossRef]
- Jaiswal, V.; Cho, Y.-I.; Lee, H.-J. Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis. Appl. Sci. 2021, 11, 226. [Google Scholar] [CrossRef]
- Sood, A.; Jaiswal, V.; Chanumolu, S.K.; Malhotra, N.; Pal, T.; Chauhan, R.S. Mining whole genomes and transcriptomes of Jatropha (Jatropha curcas) and Castor bean (Ricinus communis) for NBS-LRR genes and defense response associated transcription factors. Mol. Biol. Rep. 2014, 41, 7683–7695. [Google Scholar] [CrossRef]
- Vishambra, D.; Srivastava, M.; Dev, K.; Jaiswal, V. Subcellular localization based comparative study on radioresistant bacteria: A novel approach to mine proteins involve in radioresistance. Comput. Biol. Chem. 2017, 69, 1–9. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome analysis based on RNA-Seq in understanding pathogenic mechanisms of diseases and the immune system of fish: A comprehensive review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef] [Green Version]
- Jankowsky, J.L.; Fadale, D.J.; Anderson, J.; Xu, G.M.; Gonzales, V.; Jenkins, N.A.; Copeland, N.G.; Lee, M.K.; Younkin, L.H.; Wagner, S.L. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: Evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 2004, 13, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Xia, M.; Yan, X.; Li, D.; Wang, L.; Xu, Y.; Jin, T.; Ling, W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ. Res. 2012, 111, 967–981. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hu, Q.; Liu, X.; Zou, K.; Sarkodie, E.K.; Liu, X.; Gao, F. AllEnricher: A comprehensive gene set function enrichment tool for both model and non-model species. BMC Bioinform. 2020, 21, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.O. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Ding, H.; Lin, H.; Chen, W. AOD: The antioxidant protein database. Sci. Rep. 2017, 7, 7449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Benedetto, G.; Burgaletto, C.; Carta, A.R.; Saccone, S.; Lempereur, L.; Mulas, G.; Loreto, C.; Bernardini, R.; Cantarella, G. Beneficial effects of curtailing immune susceptibility in an Alzheimer’s disease model. J. Neuroinflamm. 2019, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sallenave, J.-M. Secretory leukocyte protease inhibitor and elafin/trappin-2: Versatile mucosal antimicrobials and regulators of immunity. Am. J. Respir. Cell Mol. Biol. 2010, 42, 635–643. [Google Scholar] [CrossRef]
- Dar, A.A.; Pradhan, T.N.; Kulkarni, D.P.; Shah, S.U.; Rao, K.V.; Chaukar, D.A.; D’Cruz, A.K.; Chiplunkar, S.V. Extracellular 2′ 5′-oligoadenylate synthetase 2 mediates T-cell receptor CD 3-ζ chain down-regulation via caspase-3 activation in oral cancer. Immunology 2016, 147, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Meffre, E.; Milili, M.; Blanco-Betancourt, C.; Antunes, H.; Nussenzweig, M.C.; Schiff, C. Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J. Clin. Investig. 2001, 108, 879–886. [Google Scholar] [CrossRef]
- Ripoll, V.M.; Meadows, N.A.; Bangert, M.; Lee, A.W.; Kadioglu, A.; Cox, R.D. Nicotinamide nucleotide transhydrogenase (NNT) acts as a novel modulator of macrophage inflammatory responses. FASEB J. 2012, 26, 3550–3562. [Google Scholar] [CrossRef]
- Ghidoni, R.; Flocco, R.; Paterlini, A.; Glionna, M.; Caruana, L.; Tonoli, E.; Binetti, G.; Benussi, L. Secretory leukocyte protease inhibitor protein regulates the penetrance of frontotemporal lobar degeneration in progranulin mutation carriers. J. Alzheimer’s Dis. 2014, 38, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Levault, K.R.; Brewer, G.J. Relative importance of redox buffers GSH and NAD (P) H in age-related neurodegeneration and Alzheimer disease-like mouse neurons. Aging Cell 2014, 13, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Doumas, S.; Kolokotronis, A.; Stefanopoulos, P. Anti-inflammatory and antimicrobial roles of secretory leukocyte protease inhibitor. Infect. Immun. 2005, 73, 1271–1274. [Google Scholar] [CrossRef] [Green Version]
- Rao, K.S.; Shen, X.; Pardue, S.; Krzywanski, D.M. Nicotinamide nucleotide transhydrogenase (NNT) regulates mitochondrial ROS and endothelial dysfunction in response to angiotensin II. Redox Biol. 2020, 36, 101650. [Google Scholar] [CrossRef]
- Schetters, S.T.; Kruijssen, L.J.; Crommentuijn, M.H.; Kalay, H.; Ochando, J.; Den Haan, J.M.; Garcia-Vallejo, J.J.; Van Kooyk, Y. Mouse DC-SIGN/CD209a as target for antigen delivery and adaptive immunity. Front. Immunol. 2018, 9, 990. [Google Scholar] [CrossRef]
- Watson, C.T.; Glanville, J.; Marasco, W.A. The individual and population genetics of antibody immunity. Trends Immunol. 2017, 38, 459–470. [Google Scholar] [CrossRef]
- Salzano, S.; Checconi, P.; Hanschmann, E.-M.; Lillig, C.H.; Bowler, L.D.; Chan, P.; Vaudry, D.; Mengozzi, M.; Coppo, L.; Sacre, S. Linkage of inflammation and oxidative stress via release of glutathionylated peroxiredoxin-2, which acts as a danger signal. Proc. Natl. Acad. Sci. USA 2014, 111, 12157–12162. [Google Scholar] [CrossRef] [Green Version]
- Szeliga, M. Peroxiredoxins in Neurodegenerative Diseases. Antioxidants 2020, 9, 1203. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Cristóvão, J.S.; Gomes, C.M. S100 Proteins in Alzheimer’s disease. Front. Neurosci. 2019, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, G.J.; Gao, Q.; Li, N.; Wang, R.t. C-type lectin-like receptor 2 and zonulin are associated with mild cognitive impairment and Alzheimer’s disease. Acta Neurol. Scand. 2020, 141, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Jeong, N.Y.; Jung, J. Therapeutic importance of hydrogen sulfide in age-associated neurodegenerative diseases. Neural Regen. Res. 2020, 15, 653. [Google Scholar] [PubMed]




| No. | Group | Sample ID | Number of Reads | Number of Good Reads (Percentage) | Overall Alignment Rate |
|---|---|---|---|---|---|
| 1 | Wt mice | VC1 | 21,622,102 | 20,743,123 (95.9%) | 93.51% |
| 2 | VC4 | 23,804,636 | 22,983,122 (96.5%) | 94.19% | |
| 3 | VC6 | 25,529,616 | 24,586,395 (96.3%) | 94.10% | |
| 4 | ADM mice | APP2 | 20,475,388 | 19,700,219 (96.2%) | 93.03% |
| 5 | APP4 | 20,315,569 | 19,541,861 (96.1%) | 91.00% | |
| 6 | APP6 | 24,571,206 | 23,772,655 (96.7%) | 93.35% | |
| 7 | ADM mice+C3G | C2 | 25,878,453 | 25,060,447 (96.8%) | 91.31% |
| 8 | C4 | 25,609,508 | 24,844,294 (97.01%) | 93.06% | |
| 9 | C6 | 24,938,833 | 24,141,244 (96.8%) | 92.61% | |
| 10 | All samples | Total: 212,745,311 Average: 23,638,367.8 | Total: 205,373,360 | Mean: 92.906% |
| Sr. No. | Gene Name | Differential Expression | Associated with Immune Function | Associated with Antioxidant Activity | Associated with AD |
|---|---|---|---|---|---|
| 1 | Slpi | Upregulated in both the comparisons | Yes | Yes | Yes |
| 2 | Oas2 | Yes | |||
| 3 | Ighv11-2 | Yes | |||
| 4 | Nnt | Yes | Yes | Yes | |
| 5 | S100a8 | Upregulated in comparison II (i.e., treatment group) | Yes | Yes | Yes |
| 6 | S100a9 | Yes | Yes | Yes | |
| 7 | Prdx2 | Yes | Yes | Yes | |
| 8 | Hp | Yes | Yes | Yes | |
| 9 | Mpst | Yes | Yes | ||
| 10 | Prxl2a | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaiswal, V.; Park, M.; Lee, H.-J. Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model. Antioxidants 2021, 10, 1435. https://doi.org/10.3390/antiox10091435
Jaiswal V, Park M, Lee H-J. Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model. Antioxidants. 2021; 10(9):1435. https://doi.org/10.3390/antiox10091435
Chicago/Turabian StyleJaiswal, Varun, Miey Park, and Hae-Jeung Lee. 2021. "Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model" Antioxidants 10, no. 9: 1435. https://doi.org/10.3390/antiox10091435
APA StyleJaiswal, V., Park, M., & Lee, H. -J. (2021). Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model. Antioxidants, 10(9), 1435. https://doi.org/10.3390/antiox10091435