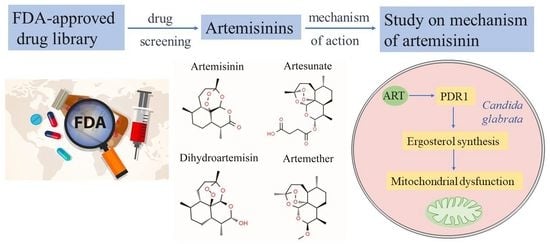
Artemisinin Targets Transcription Factor PDR1 and Impairs Candida glabrata Mitochondrial Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media and Chemicals
2.2. Screening Antifungal Drugs
2.3. Spot Assay and Growth Assay
2.4. Colony-Forming Units Counting Assay
2.5. Fungal RNA Isolation
2.6. Expression Analysis by Quantitative Real-Time PCR
2.7. Measurement of ROS Levels
2.8. Determination of Mitochondrial Membrane Potential
2.9. Mitochondrial Isolation and Membrane Viscosity Assessment
2.10. Molecular Dynamics Simulation
2.11. Statistical Analysis
3. Results
3.1. High-Throughput Screening Identifies New Antifungal Drugs
3.2. Artemisinin Is Active against Candida glabrata
3.3. Artemisinin Targets Transcription Factor PDR1
3.4. Artemisinin Effects on Ergosterol Synthesis through PDR1
3.5. Artemisinin Impairs Mitochondrial Function of Candida glabrata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.; Denning, D. Global and multi-national prevalence of fungal diseases—estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.; Pfaller, M.A.; Andes, D.R.; Diekema, D.J.; Horn, D.L.; Reboli, A.C.; Rotstein, C.; Franks, B.; Azie, N.E. Epidemiology and outcomes of invasive Candidiasis due to non-albicans species of Candida in 2496 patients: Data from the prospective antifungal therapy (PATH) registry 2004–2008. PLoS ONE 2014, 9, e101510. [Google Scholar]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant Candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216 (Suppl. S3), S445–S451. [Google Scholar] [CrossRef]
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73 (Suppl. S1), i4–i13. [Google Scholar] [CrossRef]
- Lim, C.S.Y.; Rosli, R.; Seow, H.F.; Chong, P.P. Candida and invasive candidiasis: Back to basics. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2012, 31, 21–31. [Google Scholar] [CrossRef]
- Vazquez, J.A. Therapeutic options for the management of oropharyngeal and esophageal candidiasis in HIV/AIDS patients. HIV Clin. Trials. 2000, 1, 47–59. [Google Scholar] [CrossRef]
- Campione, E.; Cosio, T.; Lanna, C.; Mazzilli, S.; Ventura, A.; Dika, E.; Gaziano, R.; Dattola, A.; Candi, E.; Bianchi, L. Predictive role of vitamin A serum concentration in psoriatic patients treated with IL-17 inhibitors to prevent skin and systemic fungal infections. J. Pharm. Sci. 2020, 144, 52–56. [Google Scholar] [CrossRef]
- Siles, S.A.; Srinivasan, A.; Pierce, C.G.; Lopez-Ribot, J.L.; Ramasubramanian, A.K. High-throughput screening of a collection of known pharmacologically active small compounds for identification of Candida albicans biofilm inhibitors. Antimicrob. Agents Chemother. 2013, 57, 3681–3687. [Google Scholar] [CrossRef]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal drugs: The current armamentarium and development of new agents. Microbiol. Spectr. 2016, 4, 19. [Google Scholar] [CrossRef]
- Fang, J.; Huang, B.; Ding, Z. Efficacy of antifungal drugs in the treatment of oral candidiasis: A Bayesian network meta-analysis. J Prosthet. Dent. 2021, 125, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R., 3rd; Cadena, J.; Patterson, T.F. Overview of antifungal agents. Clin. Chest. Med. 2009, 30, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Shekhar-Guturja, T.; Gunaherath, G.M.; Wijeratne, E.M.; Lambert, J.P.; Averette, A.F.; Lee, S.C.; Kim, T.; Bahn, Y.S.; Tripodi, F.; Ammar, R.; et al. Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nat. Chem. Biol. 2016, 12, 867–875. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug. Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef]
- LaFleur, M.D.; Lucumi, E.; Napper, A.D.; Diamond, S.L.; Lewis, K. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J. Antimicrob. Chemother. 2011, 66, 820–826. [Google Scholar] [CrossRef]
- Kaneko, Y.; Fukazawa, H.; Ohno, H.; Miyazaki, Y. Combinatory effect of fluconazole and FDA-approved drugs against Candida albicans. J. Infect. Chemother. 2013, 19, 1141–1145. [Google Scholar] [CrossRef]
- Lin, X.; Qi, Y.; Yan, D.; Liu, H.; Chen, X.; Liu, L. CgMED3 changes membrane sterol composition to help Candida glabrata tolerate low-pH stress. Appl. Env. Microbiol. 2017, 83, e00972-17. [Google Scholar] [CrossRef]
- Meisinger, C.; Pfanner, N.; Truscott, K.N. Isolation of yeast mitochondria. Methods Mol. Biol. 2006, 313, 33–39. [Google Scholar]
- Sun, C.; Li, J.; Cao, Y.; Long, G.; Zhou, B. Two distinct and competitive pathways confer the cellcidal actions of artemisinins. Microb. Cell. 2015, 2, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.J.; Kew, K.A.; Ryan, T.E.; Pennington, E.R.; Lin, C.T.; Buddo, K.A.; Fix, A.M.; Smith, C.A.; Gilliam, L.A.; Karvinen, S.; et al. 17beta-estradiol directly lowers mitochondrial membrane microviscosity and improves bioenergetic function in skeletal muscle. Cell. Metab. 2018, 27, 167–179.e167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.-H.; Chen, Q.; Yin, S.-N.; Ding, X.-W.; Zheng, Y.-C.; Zhang, Z.; Zhang, Y.-H.; Chen, F.-F.; Xu, J.-H.; Zheng, G.-W. Asymmetric reductive amination of structurally diverse ketones with ammonia using a spectrum-extended amine dehydrogenase. ACS Catal. 2021, 11, 14274–14283. [Google Scholar] [CrossRef]
- Alenquer, M.; Tenreiro, S.; Sa-Correia, I. Adaptive response to the antimalarial drug artesunate in yeast involves Pdr1p/Pdr3p-mediated transcriptional activation of the resistance determinants TPO1 and PDR5. FEMS Yeast Res. 2006, 6, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ischer, F.; Calabrese, D.; Posteraro, B.; Sanguinetti, M.; Fadda, G.; Rohde, B.; Bauser, C.; Bader, O.; Sanglard, D. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 2009, 5, e1000268. [Google Scholar] [CrossRef]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Pan, S.J.; Fan, X.; Breger, J.; Frueh, D.P.; Gulshan, K.; Li, D.K.; Mylonakis, E.; et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604–609. [Google Scholar] [CrossRef]
- Nishikawa, J.L.; Boeszoermenyi, A.; Vale-Silva, L.A.; Torelli, R.; Posteraro, B.; Sohn, Y.J.; Ji, F.; Gelev, V.; Sanglard, D.; Sanguinetti, M.; et al. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 2016, 530, 485–489. [Google Scholar] [CrossRef]
- Miller, B.R., 3rd; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-State free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Kohli, A.; Smriti; Mukhopadhyay, K.; Rattan, A.; Prasad, R. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 2002, 46, 1046–1052. [Google Scholar] [CrossRef]
- Akins, R.A. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 2005, 43, 285–318. [Google Scholar] [CrossRef]
- Dimmer, K.S.; Fritz, S.; Fuchs, F.; Messerschmitt, M.; Weinbach, N.; Neupert, W.; Westermann, B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002, 13, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 2018, 9, e01291-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Huang, L.; Li, J.; Fan, Q.; Long, Y.; Li, Y.; Zhou, B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 2010, 5, e9582. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Mo, W.; Shen, D.; Sun, L.; Wang, J.; Lu, S.; Gitschier, J.M.; Zhou, B. Yeast model uncovers dual roles of mitochondria in action of artemisinin. PLoS Genet. 2005, 1, e36. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.B.; Marella, M.; Yagi, T.; Matsuno-Yagi, A. The single subunit NADH dehydrogenase reduces generation of reactive oxygen species from complex I. FEBS Lett. 2006, 580, 6105–6108. [Google Scholar] [CrossRef]
- Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef]
- Shi, Q.; Xia, F.; Wang, Q.; Liao, F.; Guo, Q.; Xu, C.; Wang, J. Discovery and repurposing of artemisinin. Front. Med. 2022, 16, 1–9. [Google Scholar] [CrossRef]
- White, N.J. Qinghaosu (artemisinin): The price of success. Science 2008, 320, 330–334. [Google Scholar] [CrossRef]
- Kolaczkowski, M.; Kolaczkowska, A.; Stermitz, F.R. Modulation of the antifungal activity of new medicinal plant extracts active on Candida glabrata by the major transporters and regulators of the pleiotropic drug-resistance network in Saccharomyces cerevisiae. Microb. Drug Resist. 2009, 15, 11–17. [Google Scholar] [CrossRef]
- Das, S.; Czuni, L.; Balo, V.; Papp, G.; Gazdag, Z.; Papp, N.; Koszegi, T. Cytotoxic action of artemisinin and scopoletin on planktonic forms and on biofilms of Candida Species. Molecules 2020, 25, 476. [Google Scholar] [CrossRef]
- Zhou, J.; Li, J.; Cheong, I.; Liu, N.N.; Wang, H. Evaluation of artemisinin derivative artemether as a fluconazole potentiator through inhibition of Pdr5. Bioorg. Med. Chem. 2021, 44, 116293. [Google Scholar] [CrossRef]
- Zhu, C.; Liao, B.; Ye, X.; Zhou, Y.; Chen, X.; Liao, M.; Cheng, L.; Zhou, X.; Ren, B. Artemisinin elevates ergosterol levels of Candida albicans to synergise with amphotericin B against oral candidiasis. Int. J. Antimicrob. Agents 2021, 58, 106394. [Google Scholar] [CrossRef]
- Sun, C.; Cao, Y.; Zhu, P.; Zhou, B. A mitochondria-targeting artemisinin derivative with sharply increased antitumor but depressed anti-yeast and anti-malaria activities. Sci. Rep. 2017, 7, 45665. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.F.; Krol, A.A.; Sarti, K.E.; Bennett, J.E. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 2006, 50, 1384–1392. [Google Scholar] [CrossRef]
- Gautam, P.; Upadhyay, S.K.; Hassan, W.; Madan, T.; Sirdeshmukh, R.; Sundaram, C.S.; Gade, W.N.; Basir, S.F.; Singh, Y.; Sarma, P.U. Transcriptomic and proteomic profile of Aspergillus fumigatus on exposure to artemisinin. Mycopathologia 2011, 172, 331–346. [Google Scholar] [CrossRef]
- Firestone, G.L.; Sundar, S.N. Minireview: Modulation of hormone receptor signaling by dietary anticancer indoles. Mol. Endocrinol. 2009, 23, 1940–1947. [Google Scholar] [CrossRef]
- Sen, R.; Bandyopadhyay, S.; Dutta, A.; Mandal, G.; Ganguly, S.; Saha, P.; Chatterjee, M. Artemisinin triggers induction of cell-cycle arrest and apoptosis in Leishmania donovani promastigotes. J. Med. Microbiol. 2007, 56 Pt 9, 1213–1218. [Google Scholar] [CrossRef]
- Zheng, H.; Colvin, C.J.; Johnson, B.K.; Kirchhoff, P.D.; Wilson, M.; Jorgensen-Muga, K.; Larsen, S.D.; Abramovitch, R.B. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 2017, 13, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Hu, H.; Li, Y.; Wang, X.; Xu, M.; Liu, J.; Zhang, H.; Yan, Y.; Zhao, L.; Li, W.; et al. Anti-SARS-CoV-2 potential of artemisinins in vitro. ACS Infect. Dis. 2020, 6, 2524–2531. [Google Scholar] [CrossRef]





| Class | Drug | Inhibition at 12 h (%) | Inhibition at 24 h (%) |
|---|---|---|---|
| Polyenes | Amphotericin B | 68.05 | 48.47 |
| Echinocandins | Caspofungin Acetate | 81.68 | 84.28 |
| Imidazoles | Bifonazole | 62.94 | 82.78 |
| Triazoles | Fluconazole | 63.79 | 78.03 |
| Antiseptics | Benzethonium Chloride | 85.07 | 86.41 |
| Amino Acid | Binding Free Energy (kcal/mol) | Amino Acid | Binding Free Energy (kcal/mol) | Binding Free Energy Change (%) |
|---|---|---|---|---|
| R376 | −21.34 | R376W | −10.04 | 52.95 |
| Y584 | −29.71 | Y584C | −20.09 | 32.38 |
| P822 | −40.59 | P822L | −30.55 | 24.74 |
| D1082 | −25.11 | D1082G | −18.41 | 26.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, P.; Yue, C.; Zeng, X.; Chen, X. Artemisinin Targets Transcription Factor PDR1 and Impairs Candida glabrata Mitochondrial Function. Antioxidants 2022, 11, 1855. https://doi.org/10.3390/antiox11101855
Zhu P, Yue C, Zeng X, Chen X. Artemisinin Targets Transcription Factor PDR1 and Impairs Candida glabrata Mitochondrial Function. Antioxidants. 2022; 11(10):1855. https://doi.org/10.3390/antiox11101855
Chicago/Turabian StyleZhu, Pan, Chaoping Yue, Xin Zeng, and Xiulai Chen. 2022. "Artemisinin Targets Transcription Factor PDR1 and Impairs Candida glabrata Mitochondrial Function" Antioxidants 11, no. 10: 1855. https://doi.org/10.3390/antiox11101855
APA StyleZhu, P., Yue, C., Zeng, X., & Chen, X. (2022). Artemisinin Targets Transcription Factor PDR1 and Impairs Candida glabrata Mitochondrial Function. Antioxidants, 11(10), 1855. https://doi.org/10.3390/antiox11101855