Do Lipids Influence Gastrointestinal Processing: A Case Study of Major Soybean Allergen Gly m 4
Abstract
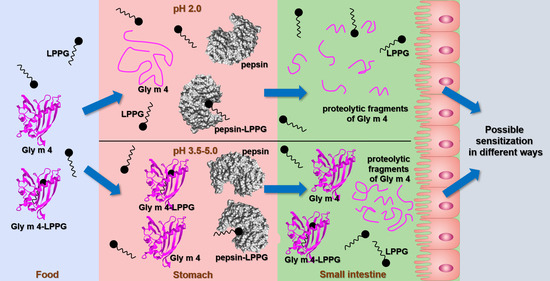
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Fluorescence Spectroscopy
2.3. Simulation of Gastric Digestion of Soybean Allergen In Vitro
2.4. CD Spectroscopy
2.5. Bioinformatic Approaches to Studying of Protein-Lipid Interactions
3. Results
3.1. Lipid-Binding Assay
3.2. Effect of pH and Lipids on Protein Secondary Structures
3.3. Proteolysis by Pepsin Mimicking Gastric Digestion under Different Conditions
3.4. Enzyme–Lipid Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rizzo, G.; Baroni, L. Soy, Soy Foods and Their Role in Vegetarian Diets. Nutrients 2018, 10, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeiger, R.S.; Sampson, H.A.; Bock, S.A.; Burks, A.W., Jr.; Harden, K.; Noone, S.; Martin, D.; Leung, S.; Wilson, G. Soy allergy in infants and children with IgE associated cow’s milk allergy. J. Pediatr. 1999, 134, 614–622. [Google Scholar] [CrossRef]
- Kosma, P.; Sjölander, S.; Landgren, E.; Borres, M.P.; Hedlin, G. Severe reactions after the intake of soy drink in birch pollen-allergic children sensitized to Gly m 4. Acta Paediatr. 2011, 100, 305–306. [Google Scholar] [CrossRef]
- Julka, S.; Kuppannan, K.; Karnoup, A.; Dielman, D.; Schafer, B.; Young, S.A. Quantification of Gly m 4 protein, a major soybean allergen, by two-dimensional liquid chromatography with ultraviolet and mass spectrometry detection. Anal. Chem. 2012, 84, 10019–10030. [Google Scholar] [CrossRef] [PubMed]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant pathogenesis-related proteins PR-10 and PR-14 as components of innate immunity system and ubiquitous allergens. Curr. Med. Chem. 2017, 24, 1772–1787. [Google Scholar] [CrossRef]
- Husslik, F.; Nürnberg, J.; Seutter von Loetzen, C.; Mews, T.; Ballmer-Weber, B.K.; Kleine-Tebbe, J.; Treudler, R.; Simon, J.C.; Randow, S.; Völker, E.; et al. The conformational IgE epitope profile of soya bean allergen Gly m 4. Clin. Exp. Allergy 2016, 46, 1484–1497. [Google Scholar] [CrossRef]
- Kleine-Tebbe, J.; Vogel, L.; Crowell, D.N.; Haustein, U.F.; Vieths, S. Severe oral allergy syndrome and anaphylactic reactions caused by a Bet v 1- related PR-10 protein in soybean, SAM22. J. Allergy Clin. Immunol. 2002, 110, 797–804. [Google Scholar] [CrossRef]
- Verma, A.K.; Kumar, S.; Das, M.; Dwivedi, P.D. A Comprehensive Review of Legume Allergy. Clin. Rev. Allergy Immunol. 2013, 45, 30–46. [Google Scholar] [CrossRef]
- Pali-Schöll, I.; Untersmayr, E.; Klems, M.; Jensen-Jarolim, E. The effect of digestion and digestibility on allergenicity of food. Nutrients 2018, 10, 1129. [Google Scholar] [CrossRef] [Green Version]
- Pekar, J.; Ret, D.; Untersmayr, E. Stability of allergens. Mol. Immunol. 2018, 100, 14–20. [Google Scholar] [CrossRef]
- Wickham, M.; Faulks, R.; Mills, C. In vitro digestion methods for assessing the effect of food structure on allergen breakdown. Mol. Nutr. Food Res. 2009, 53, 952–958. [Google Scholar] [CrossRef]
- López-Fandiño, R. Role of dietary lipids in food allergy. Crit. Rev. Food Sci. Nutr. 2020, 60, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Grzelczyk, A.; Gendaszewska-Darmach, E. Novel bioactive glycerol-based lysophospholipids: New data—New insight into their function. Biochimie 2013, 95, 667–679. [Google Scholar] [CrossRef]
- Costa, J.; Bavaro, S.L.; Benedé, S.; Diaz-Perales, A.; Bueno-Diaz, C.; Gelencser, E.; Klueber, J.; Larré, C.; Lozano-Ojalvo, D.; Lupi, R.; et al. Are Physicochemical Properties Shaping the Allergenic Potency of Plant Allergens? Clin. Rev. Allergy Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, I.V.; Finkina, E.I.; Melnikova, D.N.; Ziganshin, R.H.; Ovchinnikova, T.V. Investigation of Sensitization Potential of the Soybean Allergen Gly m 4 by Using Caco-2/Immune Cells Co-Culture Model. Nutrients 2021, 13, 2058. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer; v20.1.0.19295; Dassault Systèmes: San Diego, CA, USA, 2020. [Google Scholar]
- Clemente, T.E.; Cahoon, E.B. Soybean oil: Genetic approaches for modification of functionality and total content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Slomiany, A.; Slomiany, B.L.; Witas, H.; Zdebska, E.; Galicki, N.I.; Newman, L.J. Lipids of gastric secretion in patients with cystic fibrosis. Biochim. Biophys. Acta 1983, 750, 253–260. [Google Scholar] [CrossRef]
- Bhattacharyya, J.; Das, K.P. Molecular chaperone-like properties of an unfolded protein, alpha(s)-casein. J. Biol. Chem. 1999, 274, 15505–15509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourassa, P.; Bekale, L.; Tajmir-Riahi, H.A. Association of lipids with milk α- and β-caseins. Int. J. Biol. Macromol. 2014, 70, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Abad-Zapatero, C.; Rydel, T.J.; Erickson, J. Revised 2.3 A structure of porcine pepsin: Evidence for a flexible subdomain. Proteins 1990, 8, 62–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef]
- Suguna, K.; Padlan, E.A.; Smith, C.W.; Carlson, W.D.; Davies, D.R. Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: Implications for a mechanism of action. Proc. Natl. Acad. Sci. USA 1987, 84, 7009–7013. [Google Scholar] [CrossRef] [Green Version]
- Jacob, T.; Vogel, L.; Reuter, A.; Wangorsch, A.; Kring, C.; Mahler, V.; Wöhrl, B.M. Food Processing Does Not Abolish the Allergenicity of the Carrot Allergen Dau c 1: Influence of pH, Temperature, and the Food Matrix. Mol. Nutr. Food Res. 2020, 64, e2000334. [Google Scholar] [CrossRef]
- Mackie, A.; Mulet-Cabero, A.I.; Torcello-Gómez, A. Simulating human digestion: Developing our knowledge to create healthier and more sustainable foods. Food Funct. 2020, 11, 9397–9431. [Google Scholar] [CrossRef]
- Mennah-Govela, Y.A.; Bornhorst, G.M. Food buffering capacity: Quantification methods and its importance in digestion and health. Food Funct. 2021, 12, 543–563. [Google Scholar] [CrossRef]
- Sancho, A.I.; Wangorsch, A.; Jensen, B.M.; Watson, A.; Alexeev, Y.; Johnson, P.E.; Mackie, A.R.; Neubauer, A.; Reese, G.; Ballmer-Weber, B.; et al. Responsiveness of the major birch allergen Bet v 1 scaffold to the gastric environment: Impact on structure and allergenic activity. Mol. Nutr. Food Res. 2011, 55, 1690–1699. [Google Scholar] [CrossRef]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Matveevskaya, N.S.; Ignatova, A.A.; Toropygin, I.Y.; Ovchinnikova, T.V. Impact of Different Lipid Ligands on the Stability and IgE-Binding Capacity of the Lentil Allergen Len c 3. Biomolecules 2020, 10, 1668. [Google Scholar] [CrossRef] [PubMed]
- Gizatullina, A.K.; Finkina, E.I.; Mineev, K.S.; Melnikova, D.N.; Bogdanov, I.V.; Telezhinskaya, I.N.; Balandin, S.V.; Shenkarev, Z.O.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris. Biochem. Biophys. Res. Commun. 2013, 439, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ni, T. Probing the binding mechanisms of α-tocopherol to trypsin and pepsin using isothermal titration calorimetry, spectroscopic, and molecular modeling methods. J. Biol. Phys. 2016, 42, 415–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ignatova, A.A.; Ovchinnikova, T.V. Do Lipids Influence Gastrointestinal Processing: A Case Study of Major Soybean Allergen Gly m 4. Membranes 2021, 11, 754. https://doi.org/10.3390/membranes11100754
Finkina EI, Melnikova DN, Bogdanov IV, Ignatova AA, Ovchinnikova TV. Do Lipids Influence Gastrointestinal Processing: A Case Study of Major Soybean Allergen Gly m 4. Membranes. 2021; 11(10):754. https://doi.org/10.3390/membranes11100754
Chicago/Turabian StyleFinkina, Ekaterina I., Daria N. Melnikova, Ivan V. Bogdanov, Anastasia A. Ignatova, and Tatiana V. Ovchinnikova. 2021. "Do Lipids Influence Gastrointestinal Processing: A Case Study of Major Soybean Allergen Gly m 4" Membranes 11, no. 10: 754. https://doi.org/10.3390/membranes11100754
APA StyleFinkina, E. I., Melnikova, D. N., Bogdanov, I. V., Ignatova, A. A., & Ovchinnikova, T. V. (2021). Do Lipids Influence Gastrointestinal Processing: A Case Study of Major Soybean Allergen Gly m 4. Membranes, 11(10), 754. https://doi.org/10.3390/membranes11100754