A Study of the Interaction of a New Benzimidazole Schiff Base with Synthetic and Simulated Membrane Models of Bacterial and Mammalian Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Benzimidazole Schiff Base
2.1.1. Materials
2.1.2. Synthesis of 2-(m-aminophenyl)benzimidazole (Bz)
2.1.3. Synthesis of 4-(((3-(1H-benzo[d]imidazol-2-yl)phenyl)imino)methyl)benzene-1,3-diol (Bz-Im)
2.2. Interaction with Models of Synthetic Membranes
2.2.1. Membrane Preparation
2.2.2. Differential Scanning Calorimetry
2.3. Molecular Dynamics (MD) Studies
2.3.1. Construction of the 3D Structure of Bz-Im
2.3.2. Erythrocyte Membrane Construction
2.3.3. Construction of Gram-Negative Bacterial Membrane Models
2.3.4. Implementing Molecular Dynamics
2.3.5. Interaction Analysis
3. Results and Discussion
3.1. Schiff Base (Imine) Characterization
3.2. Model Membrane Studies
3.2.1. Thermotropic Behavior of Synthetic Model Membranes
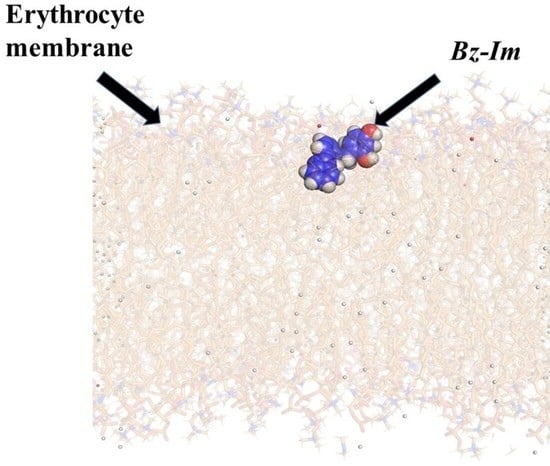
3.2.2. Analysis of Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Xu, Y.; Tillman, T.S.; Tang, P. Membranes and Drug Action, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2009; ISBN 9780123695215. [Google Scholar]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Schubert, T.; Römer, W. How synthetic membrane systems contribute to the understanding of lipid-driven endocytosis. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 2992–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef]
- Essaid, D.; Rosilio, V.; Daghildjian, K.; Solgadi, A.; Vergnaud, J.; Kasselouri, A.; Chaminade, P. Artificial plasma membrane models based on lipidomic profiling. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2725–2736. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, J.; Suhendro, D.K.; Zieleniecki, J.L.; Shapter, J.G.; Köper, I. Membrane-drug interactions studied using model membrane systems. Saudi J. Biol. Sci. 2015, 22, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oñate-Garzón, J.; Ausili, A.; Manrique-Moreno, M.; Torrecillas, A.; Aranda, F.J.; Patiño, E.; Gomez-Fernández, J.C. The increase in positively charged residues in cecropin D-like Galleria mellonella favors its interaction with membrane models that imitate bacterial membranes. Arch. Biochem. Biophys. 2017, 629, 54–62. [Google Scholar] [CrossRef]
- Rivera-Sánchez, S.P.; Agudelo-Góngora, H.A.; Oñate-Garzón, J.; Flórez-Elvira, L.J.; Correa, A.; Londoño, P.A.; Londoño-Mosquera, J.D.; Aragón-Muriel, A.; Polo-Cerón, D.; Ocampo-Ibáñez, I.D. Antibacterial Activity of a Cationic Antimicrobial Peptide against Multidrug-Resistant Gram-Negative Clinical Isolates and Their Potential Molecular Targets. Molecules 2020, 25, 5035. [Google Scholar] [CrossRef]
- Correa, W.; Manrique-Moreno, M.; Patiño, E.; Peláez-Jaramillo, C.; Kaconis, Y.; Gutsmann, T.; Garidel, P.; Heinbockel, L.; Brandenburg, K. Galleria mellonella native and analogue peptides Gm1 and Δgm1. I) Biophysical characterization of the interaction mechanisms with bacterial model membranes. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2728–2738. [Google Scholar] [CrossRef] [Green Version]
- Haralampiev, I.; Alonso de Armiño, D.J.; Luck, M.; Fischer, M.; Abel, T.; Huster, D.; Di Lella, S.; Scheidt, H.A.; Müller, P. Interaction of the small-molecule kinase inhibitors tofacitinib and lapatinib with membranes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183414. [Google Scholar] [CrossRef]
- Kaur, N.; Fischer, M.; Kumar, S.; Gahlay, G.K.; Scheidt, H.A.; Mithu, V.S. Role of cationic head-group in cytotoxicity of ionic liquids: Probing changes in bilayer architecture using solid-state NMR spectroscopy. J. Colloid Interface Sci. 2021, 581, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Lind, T.K.; Skoda, M.W.A.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Phosphatidylethanolamine and Phosphatidylglycerol by Vesicle Fusion, a Simple but Relevant Model for Bacterial Membranes. ACS Omega 2019, 4, 10687–10694. [Google Scholar] [CrossRef]
- Britt, H.M.; Prakash, A.S.; Appleby, S.; Mosely, J.A.; Sanderson, J.M. Lysis of membrane lipids promoted by small organic molecules: Reactivity depends on structure but not lipophilicity. Sci. Adv. 2020, 6, eaaz8598. [Google Scholar] [CrossRef] [Green Version]
- Dadhich, R.; Singh, A.; Menon, A.P.; Mishra, M.; Athul, C.D.; Kapoor, S. Biophysical characterization of mycobacterial model membranes and their interaction with rifabutin: Towards lipid-guided drug screening in tuberculosis. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1213–1227. [Google Scholar] [CrossRef]
- Alves, A.C.; Ribeiro, D.; Nunes, C.; Reis, S. Biophysics in cancer: The relevance of drug-membrane interaction studies. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2231–2244. [Google Scholar] [CrossRef]
- Castillo, I.; Suwalsky, M.; Gallardo, M.J.; Troncoso, V.; Sánchez-Eguía, B.N.; Santiago-Osorio, E.; Aguiñiga, I.; González-Ugarte, A.K. Structural and functional effects of benzimidazole/thioether-copper complexes with antitumor activity on cell membranes and molecular models. J. Inorg. Biochem. 2016, 156, 98–104. [Google Scholar] [CrossRef]
- Lopes-de-Campos, D.; Nunes, C.; Sarmento, B.; Jakobtorweihen, S.; Reis, S. Metronidazole within phosphatidylcholine lipid membranes: New insights to improve the design of imidazole derivatives. Eur. J. Pharm. Biopharm. 2018, 129, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Su, S.; Zhou, Q.; Tang, X.; Liu, T.; Peng, F.; He, M.; Luo, H.; Xue, W. Antibacterial and antiviral activities and action mechanism of flavonoid derivatives with a benzimidazole moiety. J. Saudi Chem. Soc. 2021, 25, 101194. [Google Scholar] [CrossRef]
- Aragón-Muriel, A.; Liscano-Martínez, Y.; Rufino-Felipe, E.; Morales-Morales, D.; Oñate-Garzón, J.; Polo-Cerón, D. Synthesis, biological evaluation and model membrane studies on metal complexes containing aromatic N,O-chelate ligands. Heliyon 2020, 6, e04126. [Google Scholar] [CrossRef] [PubMed]
- Alasmary, F.A.S.; Snelling, A.M.; Zain, M.E.; Alafeefy, A.M.; Awaad, A.S.; Karodia, N. Synthesis and evaluation of selected benzimidazole derivatives as potential antimicrobial agents. Molecules 2015, 20, 15206–15223. [Google Scholar] [CrossRef] [PubMed]
- Saluja, P.; Sharma, H.; Kaur, N.; Singh, N.; Jang, D.O. Benzimidazole-based imine-linked chemosensor: Chromogenic sensor for Mg2+ and fluorescent sensor for Cr3+. Tetrahedron 2012, 68, 2289–2293. [Google Scholar] [CrossRef]
- Dutta Gupta, S.; Revathi, B.; Mazaira, G.I.; Galigniana, M.D.; Subrahmanyam, C.V.S.; Gowrishankar, N.L.; Raghavendra, N.M. 2,4-dihydroxy benzaldehyde derived Schiff bases as small molecule Hsp90 inhibitors: Rational identification of a new anticancer lead. Bioorg. Chem. 2015, 59, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganga Raju, M.; Saritha, L.; Dutta Gupta, S.; Divya, N. Antimicrobial, Anti-Inflammatory and Anti-Parkinson’s Screening of Imine Analogues through HSP90 Inhibition. J. Chem. Pharm. Res. 2017, 9, 258–266. [Google Scholar]
- Mahmood, K.; Hashmi, W.; Ismail, H.; Mirza, B.; Twamley, B.; Akhter, Z.; Rozas, I.; Baker, R.J. Synthesis, DNA binding and antibacterial activity of metal(II) complexes of a benzimidazole Schiff base. Polyhedron 2019, 157, 326–334. [Google Scholar] [CrossRef]
- Bran̂a, M.F.; Castellano, J.M.; Yunta, M.J.R. Synthesis of benzimidazo-substituted 3-quinolinecarboxylic acids as antibacterial agents. J. Heterocycl. Chem. 1990, 27, 1177–1180. [Google Scholar] [CrossRef]
- Roopashree, B.; Gayathri, V.; Gopi, A.; Devaraju, K.S. Syntheses, characterizations, and antimicrobial activities of binuclear ruthenium(III) complexes containing 2-substituted benzimidazole derivatives. J. Coord. Chem. 2012, 65, 4023–4040. [Google Scholar] [CrossRef]
- Suman, G.R.; Bubbly, S.G.; Gudennavar, S.B.; Muthu, S.; Roopashree, B.; Gayatri, V.; Nanje Gowda, N.M. Structural investigation, spectroscopic and energy level studies of Schiff base: 2-[(3′-N-salicylidenephenyl)benzimidazole] using experimental and DFT methods. J. Mol. Struct. 2017, 1139, 247–254. [Google Scholar] [CrossRef]
- Aragón-Muriel, A.; Ausili, A.; Sánchez, K.; Rojas, A.O.E.; Londoño Mosquera, J.; Polo-Cerón, D.; Oñate-Garzón, J. Studies on the Interaction of Alyteserin 1c Peptide and Its Cationic Analogue with Model Membranes Imitating Mammalian and Bacterial Membranes. Biomolecules 2019, 9, 527. [Google Scholar] [CrossRef] [Green Version]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 2017, 70, 238–245. [Google Scholar] [CrossRef]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Huang, J.; Mackerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef] [Green Version]
- Liscano, Y.; Salamanca, C.H.; Vargas, L.; Cantor, S.; Laverde-Rojas, V.; Oñate-Garzón, J. Increases in hydrophilicity and charge on the polar face of alyteserin 1c helix change its selectivity towards gram-positive bacteria. Antibiotics 2019, 8, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrakala, M.; Sheshadri, B.S.; Nanje Gowda, N.M.; Murthy, K.G.S.; Nagasundara, K.R. Synthesis and spectral studies of 2-salicylidine-4-aminophenyl benzimidazole and its reaction with divalent Zn, Cd and Hg: Crystal structure of the cadmium bromide complex. J. Chem. Res. 2010, 576–580. [Google Scholar] [CrossRef]
- Riske, K.A.; Barroso, R.P.; Vequi-Suplicy, C.C.; Germano, R.; Henriques, V.B.; Lamy, M.T. Lipid bilayer pre-transition as the beginning of the melting process. Biochim. Biophys. Acta Biomembr. 2009, 1788, 954–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, H.W. Pretransition-ripples in bilayers of dipalmitoylphosphatidylcholine: Undulation or periodic segments? A freeze-fracture study. Biochim. Biophys. Acta Lipids Lipid Metab. 1996, 1302, 138–144. [Google Scholar] [CrossRef]
- Di Foggia, M.; Bonora, S.; Tinti, A.; Tugnoli, V. DSC and Raman study of DMPC liposomes in presence of Ibuprofen at different pH. J. Therm. Anal. Calorim. 2017, 127, 1407–1417. [Google Scholar] [CrossRef]
- Ohline, S.M.; Campbell, M.L.; Turnbull, M.T.; Kohler, S.J. Differential scanning calorimetric study of bilayer membrane phase transitions: A biophysical chemistry experiment. J. Chem. Educ. 2001, 78, 1251–1256. [Google Scholar] [CrossRef]
- Bilge, D.; Kazanci, N.; Severcan, F. Acyl chain length and charge effect on Tamoxifen-lipid model membrane interactions. J. Mol. Struct. 2013, 1040, 75–82. [Google Scholar] [CrossRef]
- Carrillo, C.; Teruel, J.A.; Aranda, F.J.; Ortiz, A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta Biomembr. 2003, 1611, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.W. Action of antimicrobial peptides: Two-state model. Biochemistry 2000, 39, 8347–8352. [Google Scholar] [CrossRef]
- Martínez, L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef] [Green Version]
- Bera, I.; Klauda, J.B. Molecular Simulations of Mixed Lipid Bilayers with Sphingomyelin, Glycerophospholipids, and Cholesterol. J. Phys. Chem. B 2017, 121, 5197–5208. [Google Scholar] [CrossRef] [PubMed]
- Björkbom, A.; Róg, T.; Kaszuba, K.; Kurita, M.; Yamaguchi, S.; Lönnfors, M.; Nyholm, T.K.M.; Vattulainen, I.; Katsumura, S.; Slotte, J.P. Effect of sphingomyelin headgroup size on molecular properties and interactions with cholesterol. Biophys. J. 2010, 99, 3300–3308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakizimana, P.; Masureel, M.; Gbaguidi, B.; Ruysschaert, J.M.; Govaerts, C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: A conserved mechanism for proton gradient sensing? J. Biol. Chem. 2008, 283, 9369–9376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blom, T.S.; Koivusalo, M.; Kuismanen, E.; Kostiainen, R.; Somerharju, P.; Ikonen, E. Mass spectrometric analysis reveals an increase in plasma membrane polyunsaturated phospholipid species upon cellular cholesterol loading. Biochemistry 2001, 40, 14635–14644. [Google Scholar] [CrossRef]
- Hauser, H.; Pascher, I.; Pearson, R.H.; Sundell, S. Preferred conformation and molecular packing of phosphatidylethanolamine and phosphatidylcholine. BBA Rev. Biomembr. 1981, 650, 21–51. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Lewis, R.N.A.H.; McElhaney, R.N. Calorimetric and spectroscopic studies of the thermotropic phase behavior of the n-saturated 1,2-diacylphosphatidylglycerols. Biophys. J. 1997, 72, 779–793. [Google Scholar] [CrossRef] [Green Version]
- Lairion, F.; Disalvo, E.A. Effect of phloretin on the dipole potential of phosphatidylcholine, phosphatidylethanolamine, and phosphatidylglycerol monolayers. Langmuir 2004, 20, 9151–9155. [Google Scholar] [CrossRef] [PubMed]






| MLV | Compound–Lipid Molar Ratio | Pretransition Temperature (°C) | Tm (°C) | ΔTm/2 (°C) | ΔH (J·g−1) |
|---|---|---|---|---|---|
| DMPC | 0:1 | 12.94 | 23.02 | 0.98 | 1.48 |
| DMPC-Compound | 1:50 | 11.58 | 22.94 | 1.09 | 1.05 |
| 1:25 | - | 22.60 | 1.14 | 0.78 | |
| 1:10 | - | 22.38 | 1.23 | 0.76 | |
| DMPC/DMPG (3:1) | 0:1 | 12.71 | 23.02 | 0.73 | 1.71 |
| DMPC/DMPG (3:1)-Compound | 1:50 | 12.49 | 22.98 | 0.90 | 1.46 |
| 1:25 | - | 22.24 | 1.08 | 1.38 | |
| 1:10 | - | 19.72 | 2.34 | 1.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aragón-Muriel, A.; Liscano, Y.; Morales-Morales, D.; Polo-Cerón, D.; Oñate-Garzón, J. A Study of the Interaction of a New Benzimidazole Schiff Base with Synthetic and Simulated Membrane Models of Bacterial and Mammalian Membranes. Membranes 2021, 11, 449. https://doi.org/10.3390/membranes11060449
Aragón-Muriel A, Liscano Y, Morales-Morales D, Polo-Cerón D, Oñate-Garzón J. A Study of the Interaction of a New Benzimidazole Schiff Base with Synthetic and Simulated Membrane Models of Bacterial and Mammalian Membranes. Membranes. 2021; 11(6):449. https://doi.org/10.3390/membranes11060449
Chicago/Turabian StyleAragón-Muriel, Alberto, Yamil Liscano, David Morales-Morales, Dorian Polo-Cerón, and Jose Oñate-Garzón. 2021. "A Study of the Interaction of a New Benzimidazole Schiff Base with Synthetic and Simulated Membrane Models of Bacterial and Mammalian Membranes" Membranes 11, no. 6: 449. https://doi.org/10.3390/membranes11060449
APA StyleAragón-Muriel, A., Liscano, Y., Morales-Morales, D., Polo-Cerón, D., & Oñate-Garzón, J. (2021). A Study of the Interaction of a New Benzimidazole Schiff Base with Synthetic and Simulated Membrane Models of Bacterial and Mammalian Membranes. Membranes, 11(6), 449. https://doi.org/10.3390/membranes11060449