Clinical Validation of Innovative Optical-Sensor-Based, Low-Cost, Rapid Diagnostic Test to Reduce Antimicrobial Resistance
Abstract
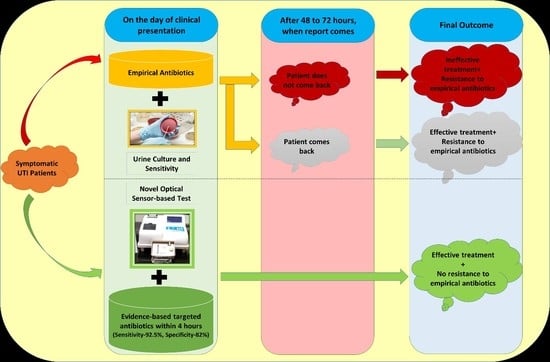
:1. Introduction
2. Materials and Methods
2.1. Study Design, Setting, and Population
2.2. Ethical Approval
2.3. Study Oversight
2.4. Test Methods for Bacterial Culture and Identification
2.5. Data Analysis
2.6. Reagents
3. Results
3.1. Study Characteristics
3.2. Test Performance
3.2.1. Diagnostic Accuracy
3.2.2. Agreement Analysis
3.3. Identification of Bacteria
3.4. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. The World Health Report 1996: Fighting DISEASE, Fostering Development; Report of the Director-General; World Health Organization: Geneva, Switzerland, 1996. [Google Scholar]
- Lal, P. Report of the National Commission on Macroeconomics and Health; Report of the National Commission on Health; Ministry of Health and Family Welfare: New Delhi, India, 2005.
- Dixit, A.; Kumar, N.; Kumar, S.; Trigun, V. Antimicrobial Resistance: Progress in the Decade since Emergence of New Delhi Metallo-β-Lactamase in India. Indian J. Community Med. 2019, 44, 4–8. [Google Scholar] [PubMed]
- Ganguly, N.K.; Arora, N.K.; Chandy, S.J.; Fairoze, M.N.; Gill, J.P.S.; Gupta, U.; Hossain, S.; Joglekar, S.; Joshi, P.C.; Kakkar, M.; et al. Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J. Med. Res. 2011, 134, 281–294. [Google Scholar] [PubMed]
- Baquero, F. Low-level antibacterial resistance: A gateway to clinical resistance. Drug Resist. Updat. 2001, 4, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Eigner, U.; Schmid, A.; Wild, U.; Bertsch, D.; Fahr, A.-M. Analysis of the comparative workflow and performance characteristics of the VITEK 2 and Phoenix systems. J. Clin. Microbiol. 2005, 43, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.S.; Cornish, N.E.; Hong, S.G.; Hemrick, K.; Herdt, C.; Moland, E.S. Comparison of Phoenix and VITEK 2 extended-spectrum-beta-lactamase detection tests for analysis of Escherichia coli and Klebsiella isolates with well-characterized beta-lactamases. J. Clin. Microbiol. 2007, 45, 2380–2384. [Google Scholar] [CrossRef]
- Dolk, F.C.K.; Pouwels, K.B.; Smith, D.R.M.; Robotham, J.V.; Smieszek, T. Antibiotics in primary care in England: Which antibiotics are prescribed and for which conditions? J. Antimicrob. Chemother. 2018, 73, ii2–ii10. [Google Scholar] [CrossRef]
- Chua, K.-P.; Fischer, M.A.; Linder, J.A. Appropriateness of outpatient antibiotic prescribing among privately insured US patients: ICD-10-CM based cross sectional study. BMJ 2019, 364, k5092. [Google Scholar] [CrossRef]
- Schmiemann, G.; Kniehl, E.; Gebhardt, K.; Matejczyk, M.M.; Hummers-Pradier, E. The diagnosis of urinary tract infection: A systematic review. Deutsches Ärzteblatt Int. 2010, 107, 361–367. [Google Scholar]
- Kim, H.Y.; Lee, S.-J.; Lee, D.S.; Yoo, J.M.; Choe, H.-S. Microbiological Characteristics of Unresolved Acute Uncomplicated Cystitis. Microb. Drug Resist. 2016, 22, 387–391. [Google Scholar] [CrossRef]
- Seifu, W.D.; Gebissa, A.D. Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect. Dis. 2018, 18, 30. [Google Scholar] [CrossRef]
- Chang, P.-C.; Hsu, Y.-C.; Hsieh, M.-L.; Huang, S.-T.; Huang, H.-C.; Chen, Y. A pilot study on Trichomonas vaginalis in women with recurrent urinary tract infections. Biomed. J. 2016, 39, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Frej-Mądrzak, M.; Teryks-Wołyniec, D.; Jama-Kmiecik, A.; Sarowska, J.; Choroszy-Król, I. Diagnosing Chlamydia Trachomatis Urinary Tract Infections—Preliminary Report. Adv. Clin. Exp. Med. 2015, 24, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.L.; Neild, G.H. Urinary tract infection. Medicine 2007, 35, 423–428. [Google Scholar] [CrossRef]
- Mathai, E.; Thomas, R.J.; Chandy, S.; Mathai, M.; Bergstrom, S. Antimicrobials for the treatment of urinary tract infection in pregnancy: Practices in southern India. Pharmacoepidemiol. Drug Saf. 2004, 13, 645–652. [Google Scholar] [CrossRef] [PubMed]
- U.S. Preventive Services Task Force. Screening for asymptomatic bacteriuria in adults: Reaffirmation recommendation statement. Am. Fam. Phys. 2010, 81, 505. [Google Scholar]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.-H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 296–310. [Google Scholar] [CrossRef]
- Gehani, M.; Kapur, S.; Bhardwaj, P.; Nag, V.; Balasubramaniam, S.; Kammili, N.; Madhuri, S. Unmet need of antenatal screening for asymptomatic bacteriuria: A risk factor for adverse outcomes of pregnancy. Indian J. Community Med. 2019, 44, 193. [Google Scholar] [CrossRef]
- Jayalakshmi, J.; Jayaram, V.S. Evaluation of various screening tests to detect asymptomatic bacteriuria in pregnant women. Indian J. Pathol. Microbiol. 2008, 51, 379–381. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Junkins, A.D.; Bartholdi, R.K. Poor Predictive Ability of Urinalysis and Microscopic Examination to Detect Urinary Tract Infection. Am. J. Clin. Pathol. 2000, 113, 709–713. [Google Scholar] [CrossRef]
- Okusanya, B.O.; Aigere, E.O.S.; Eigbefoh, J.O.; Okome, G.B.O.; Gigi, C.E. Is a chlorhexidine reaction test better than dipsticks to detect asymptomatic bacteriuria in pregnancy? J. Obstet. Gynaecol. 2014, 34, 21–24. [Google Scholar] [CrossRef]
- Shelton, S.D.; Boggess, K.A.; Kirvan, K.; Sedor, F.; Herbert, W.N. Urinary interleukin-8 with asymptomatic bacteriuria in pregnancy. Obstet. Gynecol. 2001, 97, 583–586. [Google Scholar] [PubMed]
- Mathews, J.E.; George, S.; Mathews, P.; Mathai, E.; Brahmadathan, K.N.; Seshadri, L. The Griess test: An inexpensive screening test for asymptomatic bacteriuria in pregnancy. Aust. N. Z. J. Obstet. Gynaecol. 1998, 38, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Archbald, F.J.; Verma, U.; Tejani, N.A. Screening for asymptomatic bacteriuria with Microstix. J. Reprod. Med. 1984, 29, 272–274. [Google Scholar] [PubMed]
- Bilir, F.; Akdemir, N.; Ozden, S.; Cevrioglu, A.S.; Bilir, C. Increased serum procalcitonin levels in pregnant patients with asymptomatic bacteriuria. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Waisman, Y.; Zerem, E.; Amir, L.; Mimouni, M. The Validity of the Uriscreen Test for Early Detection of Urinary Tract Infection in Children. Pediatrics 1999, 104, e41. [Google Scholar] [CrossRef]
- Stapleton, A.E.; Cox, M.E.; DiNello, R.K.; Geisberg, M.; Abbott, A.; Roberts, P.L.; Hooton, T.M. Performance of a New Rapid Immunoassay Test Kit for Point-of-Care Diagnosis of Significant Bacteriuria. J. Clin. Microbiol. 2015, 53, 2805–2809. [Google Scholar] [CrossRef]
- Nachum, R.; Arce, J.J.; Berzofsky, R.N. Gram-negative bacteriuria of pregnancy: Rapid detection by a chromogenic Limulus amoebocyte lysate assay. Obstet. Gynecol. 1986, 68, 215–219. [Google Scholar]
- Moshaver, B.; de Boer, F.; van Egmond-Kreileman, H.; Kramer, E.; Stegeman, C.; Groeneveld, P. Fast and accurate prediction of positive and negative urine cultures by flow cytometry. BMC Infect. Dis. 2016, 16, 211. [Google Scholar] [CrossRef]
- Duong, H.P.; Wissing, K.M.; Tram, N.; Mascart, G.; Lepage, P.; Ismaili, K. Accuracy of Automated Flow Cytometry-Based Leukocyte Counts to Rule Out Urinary Tract Infection in Febrile Children: A Prospective Cross-Sectional Study. J. Clin. Microbiol. 2016, 54, 2975–2981. [Google Scholar] [CrossRef]
- Hale, D.C.; Wright, D.N.; McKie, J.E.; Isenberg, H.D.; Jenkins, R.D.; Matsen, J.M. Rapid screening for bacteriuria by light scatter photometry (Autobac): A collaborative study. J. Clin. Microbiol. 1981, 13, 147–150. [Google Scholar]
- Jenkins, R.D.; Hale, D.C.; Matsen, J.M. Rapid semiautomated screening and processing of urine specimens. J. Clin. Microbiol. 1980, 11, 220–225. [Google Scholar] [PubMed]
- Tran, A.; Alby, K.; Kerr, A.; Jones, M.; Gilligan, P.H. Cost Savings Realized by Implementation of Routine Microbiological Identification by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2015, 53, 2473–2479. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, L.C.; Koelle, P.; McGee, Z.; Dewberry, L.S.; Wright, C.; Stallings, J.E.; Gates, E.; Chittur, K. Innovations in Worksite Diagnosis of Urinary Tract Infections and the Occupational Health Nurse. Workplace Health Saf. 2019, 67, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Fredborg, M.; Rosenvinge, F.S.; Spillum, E.; Kroghsbo, S.; Wang, M.; Sondergaard, T.E. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Price, C.S.; Kon, S.E.; Metzger, S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods 2014, 98, 50–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossuyt, P.M.; Irwig, L.; Craig, J.; Glasziou, P. Comparative accuracy: Assessing new tests against existing diagnostic pathways. BMJ 2006, 332, 1089–1092. [Google Scholar] [CrossRef] [Green Version]
- Kapur, S.; Gupta, S. Indigenous rapid diagnostic technology for antibiotic susceptibility testing in urinary tract infection: From bench side to bedside. BMJ Innov. 2017, 3, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Kapur, S.; Gupta, S.; Dv, P.; Pal, A.; Pant, J.; Jain, R. Rapid Sensor Based Technology: A Novel Tool for Direct Antimicrobial Susceptibility Testing in Urinary Tract Infection. Transl. Med. Biotecnol. 2014, 2, 22–28. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; ISBN1 56238-923-8. ISBN2 56238-924-6. [Google Scholar]
- Infectious Diseases Society of America; American Society of Nephrology; American Geriatric Society; Nicolle, L.E.; Bradley, S.; Colgan, R.; Rice, J.C.; Schaeffer, A.; Hooton, T.M. Infectious Diseases Society of America Guidelines for the Diagnosis and Treatment of Asymptomatic Bacteriuria in Adults. Clin. Infect. Dis. 2005, 40, 643–654. [Google Scholar] [CrossRef]
- Gehani, M.; Kapur, S. Rapid Diagnostic Test for Antimicrobial Susceptibility. Harv. Dataverse 2019, V1. [Google Scholar] [CrossRef]
- Riboli, D.F.M.; Lyra, J.C.; Silva, E.P.; Valadão, L.L.; Bentlin, M.R.; Corrente, J.E.; de Souza Rugolo, L.M.S.; de Souza, M.D.L.R. Diagnostic accuracy of semi-quantitative and quantitative culture techniques for the diagnosis of catheter-related infections in newborns and molecular typing of isolated microorganisms. BMC Infect. Dis. 2014, 14, 283. [Google Scholar] [CrossRef] [PubMed]
- Salmanzadeh-Ahrabi, S.; Kocagöz, T. Evaluation of a liquid-phase colorimetric method for rapid antibacterial susceptibility testing. Iran. J. Microbiol. 2013, 5, 259–262. [Google Scholar] [PubMed]
- Kocer, D.; Sariguzel, F.M.; Ciraci, M.Z.; Karakukcu, C.; Oz, L. Diagnostic Accuracy of a New Urinalysis System, DongJiu, for Diagnosis of Urinary Tract Infection. Ann. Clin. Lab. Sci. 2015, 45, 686–691. [Google Scholar] [PubMed]
- Huysal, K.; Budak, Y.U.; Ulusoy Karaca, A.; Aydos, M.; Kahvecioǧlu, S.; Bulut, M.; Polat, M. Diagnostic accuracy of urised automated urine microscopic sediment analyzer and dipstick parameters in predicting urine culture test results. Biochem. Medica 2012, 23, 211–217. [Google Scholar] [CrossRef]
- Kouri, T.; Fogazzi, G.; Gant, V.; Hallander, H.; Hofmann, W.; Guder, W.G. European Urinalysis Guidelines. Scand. J. Clin. Lab. Investig. Suppl. 2000, 60, 1–96. [Google Scholar] [CrossRef]
- Weir, N.-J.M.; Pattison, S.H.; Kearney, P.; Stafford, B.; Gormley, G.J.; Crockard, M.A.; Gilpin, D.F.; Tunney, M.M.; Hughes, C.M. Criteria required for an acceptable point-of-care test for UTI detection: Obtaining consensus using the Delphi technique. PLoS ONE 2018, 13, e0198595. [Google Scholar] [CrossRef] [Green Version]
- Drain, P.K.; Hyle, E.P.; Noubary, F.; Freedberg, K.A.; Wilson, D.; Bishai, W.R.; Rodriguez, W.; Bassett, I.V. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect. Dis. 2014, 14, 239–249. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | AIIMS 1 (n = 982) | Gandhi (n = 853) |
|---|---|---|
| Demographic characteristics | ||
| Age (in years) | ||
| Mean age | 43.4 | 35.7 |
| Minimum age | <1 | 1 |
| Maximum age | 95 | 90 |
| Gender | ||
| Male | 631 (64.3%) | 399 (46.8%) |
| Female | 351 (35.7%) | 454 (53.2%) |
| Referring Department | ||
| Medical Specialties | 193 | 481 |
| Surgical Specialties | 632 | 94 |
| Pediatrics | 67 | 138 |
| Obstetrics and Gynecology | 87 | 140 |
| Radio-diagnosis | 3 | 0 |
| Clinical Syndrome | ||
| Pyelonephritis | 10% | 1% |
| Cystitis | 90% | 99% |
| Contingency Tables | |||
|---|---|---|---|
| AIIMS (n = 982) | Urine Culture Positive | Urine Culture Negative | Total |
| Index test Positive | 566 (92.9%) | 72 (19.3%) | 638 |
| Index test Negative | 43 (7.1%) | 301 (80.7%) | 344 |
| Total | 609 | 373 | 982 |
| Gandhi (n = 853) | Urine Culture Positive | Urine Culture Negative | Total |
| Index test Positive | 250 (91.6%) | 100 (17.2%) | 350 |
| Index test Negative | 23 (8.4%) | 480 (82.8%) | 503 |
| Total | 273 | 580 | 853 |
| Combined (n = 1835) | Urine Culture Positive | Urine Culture Negative | Total |
| Index test Positive | 816 (92.5%) | 172 (18.0%) | 988 |
| Index test Negative | 66 (7.5%) | 781 (82.0%) | 847 |
| Total | 882 | 953 | 1835 |
| Diagnostic Accuracy | |||
| Parameters | AIIMS (n = 982) | Gandhi (n = 853) | Combined (n = 1835) |
| Sensitivity | 92.9% (95% CI: 90.6–94.8%) | 91.6% (95% CI: 87.6–94.6%) | 92.5% (95% CI: 90.6–94.2%) |
| Specificity | 80.7% (95% CI: 76.3–84.6%) | 82.8% (95% CI: 79.4–85.8%) | 82.0% (95% CI: 79.4–84.3%) |
| Agreement Analysis | |||
| Parameters | AIIMS (n = 982) | Gandhi (n = 853) | Combined (n = 1835) |
| Kappa value 1 | 0.748 | 0.692 | 0.741 |
| Standard error 1 | 0.022 | 0.025 | 0.016 |
| p value | <0.0005 | <0.0005 | <0.0005 |
| Identification of Bacteria (Single Species Identification) | |||||||
|---|---|---|---|---|---|---|---|
| AIIMS | E. coli | Enterococcus | Klebsiella | Proteus | Staphylococcus | Pseudomonas | Total |
| “n” (based on urine culture) | 324 | 104 | 79 | 4 | 1 | 48 | 560 |
| % correct identification by index test | 93% | 74% | 68% | 75% | 100% | 48% | 82% |
| Gandhi | E. coli | Enterococcus | Klebsiella | Proteus | Staphylococcus | Pseudomonas | Total |
| “n” (based on urine culture) | 92 | 22 | 100 | 8 | 22 | 8 | 252 |
| % correct identification by index test | 85% | 82% | 83% | 63% | 50% | 88% | 80% |
| AIIMS | ||||
| AIIMS Result | Total Tests Done Together | Agreement in Results | Disagreement in Results | |
| Gentamycin | R | 149 | 141 (95%) a | 8 (5%) b |
| S | 259 | 237 (92%) a | 22 (8%) c | |
| I | 7 | 0 | 7 d | |
| Amikacin | R | 65 | 65 (100%) a | 0 b |
| S | 29 | 26 (90%) a | 3 (10%) c | |
| I | 3 | 0 | 3 d | |
| Ciprofloxacin | R | 49 | 43 (88%) a | 6 (12%) b |
| S | 1 | 1 (100%) a | 0 c | |
| I | 0 | 0 | 0 d | |
| Ceftriaxone | R | 269 | 263 (98%) a | 6 (2%) b |
| S | 99 | 85 (86%) a | 14 (14%) c | |
| I | 4 | 0 | 4 d | |
| Piperacillin-Tazobactum | R | 121 | 118 (98%) a | 3 (2%) b |
| S | 286 | 266 (93%) a | 20 (7%) c | |
| I | 22 | 0 | 22 d | |
| Cefazolin | R | 34 | 24 (71%) a | 10 (29%) b |
| S | 12 | 10 (83%) a | 2 (17%) c | |
| I | 0 | 0 | 0 d | |
| Imipenem | R | 56 | 56 (100%) a | 0 b |
| S | 21 | 19 (90%) a | 2 (10%) c | |
| I | 3 | 0 | 3 d | |
| Overall | R | 743 | 710 (96%) a | 33 (4%) b |
| S | 707 | 644 (91%) a | 63 (9%) c | |
| Gandhi | ||||
| Gandhi Result | Total Tests Done Together | Agreement in Results | Disagreement in Results | |
| Gentamycin | R | 56 | 54 (96%) a | 2 (4%) b |
| S | 120 | 103 (86%) a | 17 (14%) c | |
| I | 0 | 0 | 0 d | |
| Amikacin | R | 12 | 12 (100%) a | 0 b |
| S | 27 | 23 (85%) a | 4 (15%) c | |
| I | 0 | 0 | 0 d | |
| Cefepime | R | 35 | 34 (97%) a | 1 (3%) b |
| S | 10 | 8 (80%) a | 2 (20%) c | |
| I | 0 | 0 | 0 d | |
| Piperacillin-Tazobactum | R | 13 | 12 (92%) a | 1 (8%) b |
| S | 21 | 20 (95%) a | 1 (5%) c | |
| I | 0 | 0 | 0 d | |
| Cefotaxime | R | 28 | 28 (100%) a | 0 b |
| S | 14 | 14 (100%) a | 0 c | |
| I | 0 | 0 | 0 d | |
| Levofloxacin | R | 23 | 15 (65%) a | 8 (35%) b |
| S | 8 | 6 (75%) a | 2 (25%) c | |
| I | 0 | 0 | 0 d | |
| Cefazolin | R | 56 | 50 (89%) a | 6 (11%) b |
| S | 20 | 18 (90%) a | 2 (10%) c | |
| I | 0 | 0 | 0 d | |
| Overall | R | 223 | 205 (92%) a | 18 (8%) b |
| S | 220 | 192 (87%) a | 28 (13%) c | |
| Sl. No. | Method | Sensitivity | Specificity | Detection of Bacteriuria | Identification of Bacteria | Antibiotic Susceptibility | Limitations | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | MALDI-TOF | 67% to 86% | Nearly 60% | Yes | Yes | No | Overnight incubation needed, expensive, extensive sample preparation | [18] |
| 2 | FISH | >96% | >96% | Yes | Yes | No | Requires multiple probes for all pathogens | [18] |
| 3 | PCR | 82% | 60% | Yes | Yes | No | Does not provide quantification, needs extensive initial processing and multiple probes | [18] |
| 4 | Integrated microfluidics-biosensor systems | 91% to 95% | 95% to 99% | Yes | Yes | Yes | Confounded results by urine variability and low bacterial count | [18] |
| 5 | Gram staining | 85.1% | 98.9% | Yes | No | No | [20] | |
| 6 | Dipstick with nitrite & leucocyte esterase | 53.1% | 100% | Yes | No | No | [20] | |
| 7 | Pus cell count | 42.5% | 95.5% | Yes | No | No | [20] | |
| 8 | Urine analysis and microscopy | 46.4% | 89% | Yes | No | No | [21] | |
| 9 | Chlorhexidine | 100% | 54% | Yes | No | No | [22] | |
| 10 | Interleukin-8 | 70% | 67% | Yes | No | No | [23] | |
| 11 | Griess test | 63.3% | 99.5% | Yes | No | No | [24] | |
| 12 | Serum procalcitonin level | 30% | 100% | Yes | No | No | [26] | |
| 13 | Uriscreen Test | 100% | 68.6% | Yes | No | No | [27] | |
| 14 | Antibody-based Lateral flow immunoassay | 86% | 94% | Yes | No | No | [28] | |
| 15 | Chromogenic amoebocyte lysate assay | 88.7% | 98.7% | Yes | No | No | [29] | |
| 16 | Flow cytometry-based systems | 99% | 58% | Yes | No | No | [30,31] | |
| 17 | Genetic signature identification CAPTURE assay | 100% | 90% | Yes | Yes | No | Expensive, resource intensive, infrastructure, highly skilled manpower | [35] |
| 18 | Time-lapse microscopy | 96% (agreement) | 96% (agreement) | Yes | Yes | Yes | Imprecise phenotypic identification measures in direct urine, not easy-to-use | [36,37] |
| 19 | Kirby Bauer Method | 51% | 99% | No | No | Yes | Time-consuming, resource intense, and not user-friendly | [41,42] |
| 20 | Semiquantitative culture method | 72.7% | 95.7% | Yes | Yes | Yes | Time-consuming, resource intense, and not user-friendly | [44] |
| 21 | Quantitative culture method | 59.3% | 94.4% | Yes | Yes | Yes | Time-consuming, resource intense, and not user-friendly | [44] |
| 22 | Strip-based Urinalysis DongJiu | 31.1% | 91.8% | Yes | No | No | [46] | |
| 23 | Automated Urinalysis system (URISED) | 47% | 91.1% | Yes | No | No | [47] | |
| 24 | Detection of bacteriuria by a non-culture method | 90% to 95% | 90% to 95% | Not Applicable | Not Applicable | Not Applicable | European Guidelines for Urinalysis | [48] |
| 25 | Detection of bacteriuria by a rapid non-culture method | 80% to 90% | 80% to 90% | Not Applicable | Not Applicable | Not Applicable | European Guidelines for Urinalysis | [48] |
| 26 | Index Test | 91.6 to 93.8% | 80.7 to 96.7% | Yes | Yes | Yes | User-friendly, portable, affordable, rapid-fastest available, no special training required | Data from current study described above |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapur, S.; Gehani, M.; Kammili, N.; Bhardwaj, P.; Nag, V.; Devara, S.M.; Sharad, S. Clinical Validation of Innovative Optical-Sensor-Based, Low-Cost, Rapid Diagnostic Test to Reduce Antimicrobial Resistance. J. Clin. Med. 2019, 8, 2098. https://doi.org/10.3390/jcm8122098
Kapur S, Gehani M, Kammili N, Bhardwaj P, Nag V, Devara SM, Sharad S. Clinical Validation of Innovative Optical-Sensor-Based, Low-Cost, Rapid Diagnostic Test to Reduce Antimicrobial Resistance. Journal of Clinical Medicine. 2019; 8(12):2098. https://doi.org/10.3390/jcm8122098
Chicago/Turabian StyleKapur, Suman, Manish Gehani, Nagamani Kammili, Pankaj Bhardwaj, Vijayalakshmi Nag, Sudha M. Devara, and Shashwat Sharad. 2019. "Clinical Validation of Innovative Optical-Sensor-Based, Low-Cost, Rapid Diagnostic Test to Reduce Antimicrobial Resistance" Journal of Clinical Medicine 8, no. 12: 2098. https://doi.org/10.3390/jcm8122098
APA StyleKapur, S., Gehani, M., Kammili, N., Bhardwaj, P., Nag, V., Devara, S. M., & Sharad, S. (2019). Clinical Validation of Innovative Optical-Sensor-Based, Low-Cost, Rapid Diagnostic Test to Reduce Antimicrobial Resistance. Journal of Clinical Medicine, 8(12), 2098. https://doi.org/10.3390/jcm8122098