A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
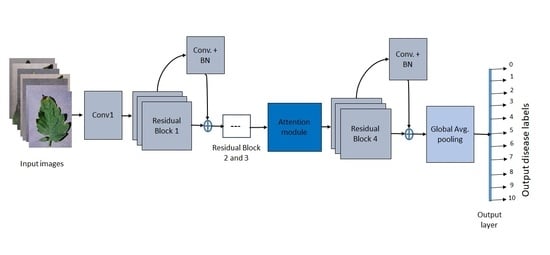
2.2. Lightweight Attention-Based Network Design
2.2.1. Convolutional Block Attention Module (CBAM)
2.2.2. Squeeze-and-Excitation (SE) Attention Module
2.2.3. Self-Attention (SA) Module
2.2.4. Dual Attention (DA) Module
2.3. Network Training and Evaluation
3. Results
3.1. Training, Validation, and Testing Accuracy of the Models
3.2. Network Parameters and Efficiency
4. Discussion
4.1. Tomato Disease Detection
4.2. Performance Evaluation of the Models
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations [FAO]. Fao Publications Catalogue 2019; FAO: Rome, Italy, 2019; p. 114. [Google Scholar]
- Liu, J.; Wang, X. Tomato Diseases and Pests Detection Based on Improved Yolo V3 Convolutional Neural Network. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017, 2917536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO. Climate-Related Transboundary Pests and Diseases; FAO: Rome, Italy, 2008; p. 59. [Google Scholar]
- Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to digit recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Gomes, J.F.S.; Leta, F.R. Applications of computer vision techniques in the agriculture and food industry: A review. Eur. Food Res. Technol. 2012, 235, 989–1000. [Google Scholar] [CrossRef]
- Garcia, J.; Barbedo, A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Bhujel, A.; Khan, F.; Basak, J.K.; Jaihuni, M.; Sihalath, T.; Moon, B.E.; Park, J.; Kim, H.T. Detection of gray mold disease and its severity on strawberry using deep learning networks. J. Plant Dis. Prot. 2022. (Accepted). [Google Scholar]
- Toda, Y.; Okura, F. How Convolutional Neural Networks Diagnose Plant Disease. Plant Phenomics 2019, 2019, 9237136. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Mnih, V.; Heess, N.; Graves, A.; Kavukcuoglu, K. Recurrent models of visual attention. In Proceedings of the Advances in Neural Information Processing Systems: Annual Conference on Neural Information Processing Systems 2014, Montreal, QC, Canada, 8–13 December 2014; Volume 3, pp. 2204–2212. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; Volume 11211, pp. 3–19. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Li, M. Crop leaf disease recognition based on Self-Attention convolutional neural network. Comput. Electron. Agric. 2020, 172, 105341. [Google Scholar] [CrossRef]
- Lee, S.H.; Goëau, H.; Bonnet, P.; Joly, A. Attention-Based Recurrent Neural Network for Plant Disease Classification. Front. Plant Sci. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Zhao, S.; Peng, Y.; Liu, J.; Wu, S. Tomato leaf disease diagnosis based on improved convolution neural network by attention module. Agriculture 2021, 11, 651. [Google Scholar] [CrossRef]
- Yilma, G.; Belay, S.; Kumie, G.; Assefa, M.; Ayalew, M.; Oluwasanmi, A.; Qin, Z. Attention augmented residual network for tomato disease detection and classification. Turkish J. Electr. Eng. Comput. Sci. 2021, 29, 2869–2885. [Google Scholar] [CrossRef]
- Yang, G.; He, Y.; Yang, Y.; Xu, B. Fine-Grained Image Classification for Crop Disease Based on Attention Mechanism. Front. Plant Sci. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep Learning for Tomato Diseases: Classification and Symptoms Visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Rangarajan, A.K.; Purushothaman, R.; Ramesh, A. Tomato crop disease classification using pre-trained deep learning algorithm. Procedia Comput. Sci. 2018, 133, 1040–1047. [Google Scholar] [CrossRef]
- Tm, P.; Pranathi, A.; Saiashritha, K.; Chittaragi, N.B.; Koolagudi, S.G. Tomato Leaf Disease Detection Using Convolutional Neural Networks. In Proceedings of the 2018 11th International Conference on Contemporary Computing (IC3), Noida, India, 2–4 August 2018. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2011–2023. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.P.; Salathe, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Koning, S.; Greeven, C.; Postma, E. Reducing Artificial Neural Network Complexity: A Case Study on Exoplanet Detection Related Work on Parameter Reduction. arXiv 2015, arXiv:1902.10385. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multi-Class Classification: An Overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Vu, T.; Wen, E.; Nehoran, R. How Not to Give a FLOP: Combining Regularization and Pruning for Efficient Inference. arXiv 2020, arXiv:2003.13593. [Google Scholar]
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 106279. [Google Scholar] [CrossRef]
- Maeda-Gutiérrez, V.; Galván-Tejada, C.E.; Zanella-Calzada, L.A.; Celaya-Padilla, J.M.; Galván-Tejada, J.I.; Gamboa-Rosales, H.; Luna-García, H.; Magallanes-Quintanar, R.; Guerrero Méndez, C.A.; Olvera-Olvera, C.A. Comparison of Convolutional Neural Network Architectures for Classification of Tomato Plant Diseases. Appl. Sci. 2020, 10, 1245. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X. Can deep learning identify tomato leaf disease? Adv. Multimed. 2018, 2018, 6710865. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, N.K.; Gautam, V.; Anand, A.; Aljahdali, H.M.; Villar, S.G.; Anand, D.; Goyal, N.; Kadry, S. Early Detection and Classification of Tomato Leaf Disease Using High-Performance Deep Neural Network. Sensors 2021, 21, 7987. [Google Scholar] [CrossRef]
- Gonzalez-huitron, V.; Le, A.; Amabilis-sosa, L.E.; Ramírez-pereda, B.; Rodriguez, H. Disease detection in tomato leaves via CNN with lightweight architectures implemented in Raspberry Pi 4. Comput. Electron. Agric. 2021, 181, 105951. [Google Scholar] [CrossRef]
- Ramamurthy, K.; Hariharan, M.; Sundar, A.; Mathikshara, P.; Johnson, A.; Menaka, R. Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. J. 2019, 86, 105933. [Google Scholar] [CrossRef]











| Class Label | Disease Common Name | Scientific Name | Images (No.) | Source (%) | |
|---|---|---|---|---|---|
| Public | Field | ||||
| 0 | Bacterial spot | Xanthomonas campestris pv. vesicatoria | 2127 | 100 | |
| 1 | Early blight | Alternaria solani | 1000 | 100 | |
| 2 | Fusarium wilt | Fusarium oxysporum f.sp. lycopersici | 1350 | - | 100 |
| 3 | Healthy | - | 1591 | 100 | |
| 4 | Late blight | Phytophthora infestans | 1909 | 100 | |
| 5 | Leaf mold | Fulvia fulva | 952 | 100 | |
| 6 | Mosaic virus | Tomato mosaic virus | 373 | 100 | |
| 7 | Septoria leaf spot | Septoria lycopersici | 1771 | 100 | |
| 8 | Spider mites | Tetranychus urticae | 1676 | 100 | |
| 9 | Target spot | Corynespora cassiicola | 1404 | 100 | |
| 10 | Yellow leaf curl virus | Begomovirus (Fam. Geminiviridae) | 5357 | 100 | |
| Total | 19,510 | ||||
| Block | Sub-Block | Layer | Kernel Size, Stride and Number | Output Shape |
|---|---|---|---|---|
| Input image | Input | 256 × 256 × 3 | ||
| Conv1 | Convolutional | 7 × 7, 2, 16 | 128 × 128 × 16 | |
| Residual block1 | Convolutional block | ” | 1 × 1, 2, 16 | 63 × 63 × 64 |
| ” | 3 × 3, 1, 16 | |||
| ” | 1 × 1, 1, 64 | |||
| Shortcut | ” | 1 × 1, 2, 64 | ||
| Residual block2 | Convolutional block | ” | 1 × 1, 2, 32 | 32 × 32 × 128 |
| ” | 3 × 3, 1, 32 | |||
| ” | 1 × 1, 1, 128 | |||
| Shortcut | ” | 1 × 1, 2, 128 | ||
| Identity block | ” | 1 × 1, 1, 32 | ||
| ” | 3 × 3, 1, 32 | |||
| ” | 1 × 1, 1, 128 | |||
| Residual block3 | Convolutional block | ” | 1 × 1, 2, 64 | 16 × 16 × 256 |
| ” | 3 × 3, 1, 64 | |||
| ” | 1 × 1, 1, 256 | |||
| Shortcut | ” | 1 × 1, 2, 256 | ||
| Identity block | ” | 1 × 1, 1, 64 | ||
| ” | 3 × 3, 1, 64 | |||
| ” | 1 × 1, 1, 256 | |||
| Residual block4 | Convolutional block | ” | 1 × 1, 2, 256 | 8 × 8 × 1024 |
| ” | 3 × 3, 1, 256 | |||
| ” | 1 × 1, 1, 1024 | |||
| Shortcut | ” | 1 × 1, 2, 1024 | ||
| Global average pooling | Global average pooling | 1024 | ||
| Dense | Output | 11 |
| Particular | Description |
|---|---|
| Batch size | 32 |
| Optimizer | Adam |
| Learning rate | 0.001 (default) |
| Epoch | 100 |
| Data augmentation: | |
| Image zooming | 0.2 |
| Vertical shearing | 0.2 |
| Horizontal shearing | 0.2 |
| Vertical flip | True |
| Horizontal flip | True |
| Vertical shift | 0.2 |
| Horizontal shift | 0.2 |
| Image rotation | 45° |
| Class Name | Training | Validation * | Testing * | |
|---|---|---|---|---|
| Original Count | After Augmentation | |||
| Bacterial spot | 1701 | 15,309 | 212 | 214 |
| Early blight | 800 | 7200 | 100 | 100 |
| Fusarium wilt | 1080 | 9720 | 135 | 135 |
| Healthy | 1272 | 11,448 | 159 | 160 |
| Late blight | 1527 | 13,743 | 190 | 192 |
| Leaf mold | 761 | 6849 | 95 | 96 |
| Mosaic virus | 298 | 2682 | 37 | 38 |
| Septoria leaf spot | 1416 | 12,744 | 177 | 178 |
| Spider mites | 1340 | 12,060 | 167 | 169 |
| Target spot | 1123 | 10,107 | 140 | 141 |
| Yellow leaf curl virus | 4285 | 38,565 | 535 | 537 |
| Total count | 15,603 | 140,427 | 1947 | 1960 |
| Model | Training | Validation | ||
|---|---|---|---|---|
| Loss | Accuracy | Loss | Accuracy | |
| lw_resnet20 | 0.0155 | 0.9954 | 0.0194 | 0.9936 |
| lw_resnet20_cbam | 0.0186 | 0.9942 | 0.0155 | 0.9951 |
| lw_resnet20_se | 0.0169 | 0.9942 | 0.0300 | 0.9905 |
| lw_resnet20_sa | 0.0205 | 0.9938 | 0.0198 | 0.9947 |
| lw_resnet20_da | 0.0205 | 0.9925 | 0.0191 | 0.9947 |
| * Class Label | lw_resnet20 | lw_resnet20_cbam | lw_resnet20_se | lw_resnet20_sa | lw_resnet20_da | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| prec. | rec. | F1 Score | prec. | rec. | F1 Score | prec. | rec. | F1 Score | prec. | rec. | F1 Score | prec. | rec. | F1 Score | |
| 0 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1 | 0.96 | 0.97 | 0.97 | 1.00 | 0.99 | 0.99 | 0.97 | 0.98 | 0.98 | 1.00 | 0.97 | 0.98 | 0.95 | 0.99 | 0.97 |
| 2 | 0.99 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 |
| 3 | 1.00 | 0.97 | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | 0.97 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 0.97 | 0.99 |
| 4 | 0.99 | 0.95 | 0.97 | 0.99 | 1.00 | 0.99 | 0.96 | 1.00 | 0.98 | 0.99 | 1.00 | 0.99 | 0.98 | 0.96 | 0.97 |
| 5 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 0.99 | 0.99 | 1.00 | 0.98 | 0.99 | 0.99 | 0.98 | 0.98 |
| 6 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 0.96 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 0.99 | 0.98 | 1.00 | 0.99 | 0.97 | 1.00 | 0.99 |
| 8 | 1.00 | 0.97 | 0.98 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 0.95 | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | 0.99 |
| 9 | 0.95 | 0.96 | 0.96 | 0.99 | 0.99 | 0.99 | 0.97 | 0.93 | 0.95 | 0.97 | 1.00 | 0.99 | 0.99 | 0.98 | 0.98 |
| 10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 |
| Avg. acc. | 0.9858 | 0.9969 | 0.9885 | 0.9932 | 0.9890 | ||||||||||
| Model | Network Parameters | Training Time (h:m:s) | Test Time per Image (ms) | Size on Disk (MB) | GFLOPs | Average Accuracy (%) |
|---|---|---|---|---|---|---|
| lw_resnet20 | 1,424,043 | 3:42:47 | 0.795 | 16.6 | 0.439 | 98.58 |
| lw_resnet20_cbam | 1,440,813 | 3:45:16 | 0.914 | 16.8 | 0.440 | 99.69 |
| lw_resnet20_se | 1,440,715 | 3:43:24 | 0.927 | 16.8 | 0.440 | 98.85 |
| lw_resnet20_sa | 1,572,075 | 3:44:14 | 0.961 | 18.3 | 0.553 | 99.32 |
| lw_resnet20_da | 1,505,963 | 3:46:27 | 0.984 | 17.6 | 0.587 | 98.90 |
| Standard ResNet50 | 23,610,251 | 3:45:51 | 1.591 | 270 | 10.10 | 98.74 |
| Reference | Deep CNN Architecture | Datasets | Accuracy (%) | Summary |
|---|---|---|---|---|
| [2] | Improved YOLOV3 | 12 classes of tomato leaf images | 92.39 | Applied feature fusion technique for tiny disease spot detection. |
| [18] | ResNet50 with squeeze-and-excitation (SE) attention module | 10 classes of tomato leaf and 4 classes of grape leaf images | 96.81 for tomato and 99.24 for grape datasets | SE attention module was implemented into a generic ResNet50 model. |
| [19] | Attention augmented ResNet (AAR) | 10 classes of tomato leaf images | 98.91 | Synthetically generates tomato leaf images to increase training datasets. |
| [22] | AlexNet and VGG16 (pretrained) | 7 classes of tomato leaf images | 97.49 for AlexNet and 97.29 for VGG16 model | Applied transfer learning strategy to swallow and medium deep models. |
| [32] | DenseNet121 (pretrained) | 5, 7, and 10 classes of tomato leaf images | 99.51, 98.65, and 97.11 for 5, 7, and 10 classes, respectively | Used transfer learning with original and synthetically generated images. |
| [33] | AlexNet, GoogleNet, Inception V3, ResNet18, and ResNet50 | 10 classes of tomato leaf images | 98.93, 99.39, 98.65, 99.06, and 99.15, respectively | Compared the performances of some standard models. |
| [34] | AlexNet, GoogleNet, and ResNet50 | 9 classes of tomato leaf images | 95.83, 95.66, and 96.51, respectively | Used only the diseased images of tomato leaves with generic model. |
| [35] | Customized 11 layer CNN | 10 classes of tomato leaf images | 98.49 | Used a swallow network with 3000 images only. |
| [36] | MobileNetV2, NASNetMobile, Xception, MobileNet V3 (pretrained) | ” | 75, 84, 100, and 98, respectively | Used lightweight generic models to deploy in Raspberry Pi. |
| [37] | Attention embedded ResNet | 4 classes of tomato leaf images | 98 | Applied an attention-based swallow ResNet model for small datasets. |
| Our model | Lightweight CBAM attention module-based ResNet20 (lw_resnet20_cbam) | 11 classes of tomato leaf images | 99.69 | Reduced the network parameters and complexity of backbone network and improved performance using an effective attention module. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhujel, A.; Kim, N.-E.; Arulmozhi, E.; Basak, J.K.; Kim, H.-T. A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture 2022, 12, 228. https://doi.org/10.3390/agriculture12020228
Bhujel A, Kim N-E, Arulmozhi E, Basak JK, Kim H-T. A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture. 2022; 12(2):228. https://doi.org/10.3390/agriculture12020228
Chicago/Turabian StyleBhujel, Anil, Na-Eun Kim, Elanchezhian Arulmozhi, Jayanta Kumar Basak, and Hyeon-Tae Kim. 2022. "A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification" Agriculture 12, no. 2: 228. https://doi.org/10.3390/agriculture12020228
APA StyleBhujel, A., Kim, N.-E., Arulmozhi, E., Basak, J. K., & Kim, H.-T. (2022). A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture, 12(2), 228. https://doi.org/10.3390/agriculture12020228