Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Positively Charged Silver Colloids and SERS Measurements
2.3. Instrumentation
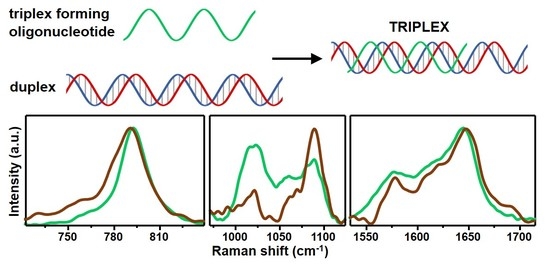
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tateishi-Karimata, H.; Sugimoto, N. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. 2020, 56, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wang, G.; Vasquez, K.M. DNA triple helices: Biological consequences and therapeutic potential. Biochimie 2008, 90, 1117–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, P.P.; Glazer, P.M. Triplex DNA: Fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997, 75, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Cecconello, A.; Idili, A.; Ricci, F.; Willner, I. Triplex DNA nanostructures: From basic properties to applications. Angew. Chem. Int. Edit. 2017, 56, 15210–15233. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Rusling, D.A. Triplex-forming oligonucleotides: A third strand for DNA nanotechnology. Nucleic Acids Res. 2017, 46, 1021–1037. [Google Scholar] [CrossRef] [Green Version]
- Sau, S.P.; Kumar, P.; Sharma, P.K.; Hrdlicka, P.J. Fluorescent intercalator displacement replacement (FIDR) assay: Determination of relative thermodynamic and kinetic parameters in triplex formation—A case study using triplex-forming LNAs. Nucleic Acids Res. 2012, 40, e162. [Google Scholar] [CrossRef] [Green Version]
- Liquier, J.; Gouyette, C.; Huynh-Dinh, T.; Taillandier, E. Raman spectroscopy of nucleic acid triple helices. J. Raman Spectrosc. 1999, 30, 657–666. [Google Scholar] [CrossRef]
- Klener, J.; Štěpánek, J. UV resonance Raman study of PolyA and PolyU complexes: Mg2+-induced formation of PolyU·PolyA·PolyU triplexes. Vib. Spectrosc. 2015, 81, 32–39. [Google Scholar] [CrossRef]
- Gfrörer, A.; Schnetter, M.E.; Wolfrum, J.; Greulich, K.O. Double and triple helices of nucleic acid polymers, studied by UV-resonance Raman spectroscopy. Ber. Bunsenges. Phys. Chem. 1993, 97, 155–162. [Google Scholar] [CrossRef]
- O’Connor, T.; Bina, M. The structure of triple helical Poly(U)·Poly(A)·Poly(U) studied by Raman spectroscopy. J. Biomol. Struct. Dyn. 1984, 2, 615–625. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-enhanced Raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Edit. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.X.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, S.-C.; Hu, S.; Yan, S.; Ren, B. Fundamental understanding and applications of plasmon-enhanced Raman spectroscopy. Nat. Rev. Phys. 2020, 2, 253–271. [Google Scholar] [CrossRef]
- Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Direct surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids: From fundamental studies to real-life applications. Chem. Soc. Rev. 2018, 47, 4909–4923. [Google Scholar] [CrossRef]
- Qi, G.; Wang, D.; Li, C.; Ma, K.; Zhang, Y.; Jin, Y. Plasmonic SERS Au nanosunflowers for sensitive and label-free diagnosis of DNA base damage in stimulus-induced cell apoptosis. Anal. Chem. 2020, 92, 11755–11762. [Google Scholar] [CrossRef]
- Li, Y.; Gao, T.; Xu, G.; Xiang, X.; Han, X.; Zhao, B.; Guo, X. Base-Pair Contents and sequences of DNA double helices differentiated by surface-enhanced Raman spectroscopy. J. Phys. Chem. Lett. 2019, 10, 3013–3018. [Google Scholar] [CrossRef]
- Li, Y.; Han, X.; Zhou, S.; Yan, Y.; Xiang, X.; Zhao, B.; Guo, X. Structural features of DNA G-quadruplexes revealed by surface-enhanced Raman spectroscopy. J. Phys. Chem. Lett. 2018, 9, 3245–3252. [Google Scholar] [CrossRef] [PubMed]
- Sagar, D.M.; Korshoj, L.E.; Hanson, K.B.; Chowdhury, P.P.; Otoupal, P.B.; Chatterjee, A.; Nagpal, P. High-throughput block optical DNA sequence identification. Small 2018, 14, 1703165. [Google Scholar] [CrossRef] [PubMed]
- Morla-Folch, J.; Gisbert-Quilis, P.; Masetti, M.; Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Conformational SERS classification of K-Ras point mutations for cancer diagnostics. Angew. Chem. Int. Edit. 2017, 56, 2381–2385. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.M.; Wang, J.; Richards, R.S.; Farrell, A.; Yaxley, J.W.; Samaratunga, H.; Teloken, P.E.; Roberts, M.J.; Coughlin, G.D.; Lavin, M.F.; et al. Design and clinical verification of surface-enhanced Raman spectroscopy diagnostic technology for individual cancer risk prediction. ACS Nano 2018, 12, 8362–8371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Koo, K.M.; Wee, E.J.H.; Wang, Y.; Trau, M. A nanoplasmonic label-free surface-enhanced Raman scattering strategy for non-invasive cancer genetic subtyping in patient samples. Nanoscale 2017, 9, 3496–3503. [Google Scholar] [CrossRef] [PubMed]
- Dick, S.; Bell, S.E.J.; Alexander, K.J.; O’Neil, I.A.; Cosstick, R. SERS and SERRS detection of the DNA lesion 8-Nitroguanine: A self-labeling modification. Chem. Eur. J. 2017, 23, 10663–10669. [Google Scholar] [CrossRef] [PubMed]
- Morla-Folch, J.; Alvarez-Puebla, R.A.; Guerrini, L. Direct quantification of DNA base composition by surface-enhanced Raman scattering spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3037–3041. [Google Scholar] [CrossRef] [PubMed]
- Morla-Folch, J.; Xie, H.-N.; Gisbert-Quilis, P.; Gómez-de Pedro, S.; Pazos-Perez, N.; Alvarez-Puebla, R.A.; Guerrini, L. Ultrasensitive direct quantification of nucleobase modifications in DNA by surface-enhanced Raman scattering: The case of cytosine. Angew. Chem. Int. Edit. 2015, 54, 13650–13654. [Google Scholar] [CrossRef] [PubMed]
- Torres-Nunez, A.; Faulds, K.; Graham, D.; Alvarez-Puebla, R.A.; Guerrini, L. Silver colloids as plasmonic substrates for direct label-free surface-enhanced Raman scattering analysis of DNA. Analyst 2016, 141, 5170–5180. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Bai, C.; Wei, Y.; Tang, Y. Surface-enhanced Fourier transform Raman scattering study on the adsorption structure of an RNA triple helix at a silver electrode. Appl. Spectrosc. 1996, 50, 48–52. [Google Scholar] [CrossRef]
- Fang, Y.; Wei, Y.; Bai, C.; Kan, L.S. Surface-enhanced Fourier transform Raman scattering from a DNA triple helix poly[dA]·2poly[dT] at a silver electrode: Beyond the short-range mechanism. J. Phys. Chem. 1996, 100, 17410–17413. [Google Scholar] [CrossRef]
- Sanchez-Cortes, S.; Garcia-Ramos, J.V. Anomalous Raman bands appearing in surface-enhanced Raman spectra. J. Raman Spectrosc. 1998, 29, 365–371. [Google Scholar] [CrossRef]
- Guerrini, L.; Krpetić, Ž.; van Lierop, D.; Alvarez-Puebla, R.A.; Graham, D. Direct surface-enhanced Raman scattering analysis of DNA duplexes. Angew. Chem. Int. Edit. 2015, 54, 1144–1148. [Google Scholar] [CrossRef]
- Van Lierop, D.; Krpetic, Z.; Guerrini, L.; Larmour, I.A.; Dougan, J.A.; Faulds, K.; Graham, D. Positively charged silver nanoparticles and their effect on surface-enhanced Raman scattering of dye-labelled oligonucleotides. Chem. Commun. 2012, 48, 8192–8194. [Google Scholar] [CrossRef]
- Kunkler, C.N.; Hulewicz, J.P.; Hickman, S.C.; Wang, M.C.; McCown, P.J.; Brown, J.A. Stability of an RNA•DNA–DNA triple helix depends on base triplet composition and length of the RNA third strand. Nucleic Acids Res. 2019, 47, 7213–7222. [Google Scholar] [CrossRef] [PubMed]
- Gisbert-Quilis, P.; Masetti, M.; Morla-Folch, J.; Fitzgerald, J.M.; Pazos-Perez, N.; Garcia-Rico, E.; Giannini, V.; Alvarez-Puebla, R.A.; Guerrini, L. The structure of short and genomic DNA at the interparticle junctions of Cationic nanoparticles. Adv. Mater. Interfaces 2017, 4, 1700724. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrini, L.; Alvarez-Puebla, R.A. Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy. Nanomaterials 2021, 11, 326. https://doi.org/10.3390/nano11020326
Guerrini L, Alvarez-Puebla RA. Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy. Nanomaterials. 2021; 11(2):326. https://doi.org/10.3390/nano11020326
Chicago/Turabian StyleGuerrini, Luca, and Ramon A. Alvarez-Puebla. 2021. "Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy" Nanomaterials 11, no. 2: 326. https://doi.org/10.3390/nano11020326
APA StyleGuerrini, L., & Alvarez-Puebla, R. A. (2021). Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy. Nanomaterials, 11(2), 326. https://doi.org/10.3390/nano11020326