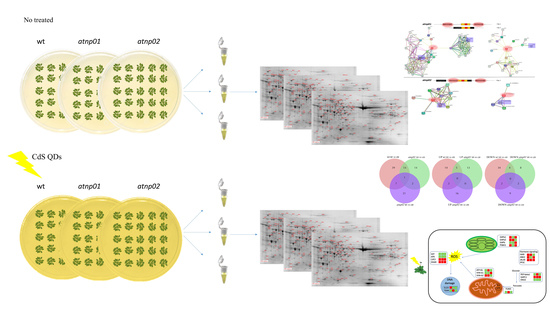
Comparative Analysis of Proteins Regulated during Cadmium Sulfide Quantum Dots Response in Arabidopsis thaliana Wild Type and Tolerant Mutants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Synthesis and Characterization of the CdS QDs
2.3. Seed Germination, Growth, and Treatments
2.4. Protein Extraction and Quantification
2.5. 2D Gel Electrophoresis
2.6. MALDI-TOF/TOF Mass Spectrometry
2.7. Data Mining and Analysis
3. Results and Discussion
3.1. A. Thaliana Proteome after CdS QDs Treatment
3.2. Ontology Analysis of the Identified Proteins
3.3. Pathways Analysis in Response to CdS QDs Treatment
3.4. Network Analysis
3.5. Comparison between 2D-PAGE, Pf2D, and Transcriptomics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cota-Ruiz, K.; Delgado-Rios, M.; Martínez-Martínez, A.; Núñez-Gastelum, J.A.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Current findings on terrestrial plants—Engineered nanomaterial interactions: Are plants capable of phytoremediating nanomaterials from soil? Curr. Opin. Environ. Sci. Health 2018, 6, 9–15. [Google Scholar] [CrossRef]
- Hatami, M.; Kariman, K.; Ghorbanpour, M. Engineered nanomaterial-mediated changes in the metabolism of terrestrial plants. Sci. Total. Environ. 2016, 571, 275–291. [Google Scholar] [CrossRef]
- Alivisatos, A.P. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science 1996, 271, 933–937. [Google Scholar] [CrossRef] [Green Version]
- Matea, C.T.; Mocan, T.; Tabaran, F.; Pop, T.; Mosteanu, O.; Puia, C.; Iancu, C.; Mocan, L. Quantum dots in imaging, drug delivery and sensor applications. Int. J. Nanomed. 2017, 12, 5421–5431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, C.; Gong, Y. Current Status and Future Prospects of Semiconductor Quantum Dots in Botany. J. Agric. Food Chem. 2019, 67, 7561–7568. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, P.; Kumar, A.; Kumar, S.; Chaudhary, R.K.; Gulyás, B. Nanoparticles in practice for molecular-imaging applications: An overview. Acta Biomater. 2016, 41, 1–16. [Google Scholar] [CrossRef]
- Wang, Z.-G.; Liu, S.-L.; Hu, Y.-J.; Tian, Z.-Q.; Hu, B.; Zhang, Z.-L.; Pang, D.-W. Dissecting the Factors Affecting the Fluorescence Stability of Quantum Dots in Live Cells. ACS Appl. Mater. Interfaces 2016, 8, 8401–8408. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Wang, P.; Xu, L.; Wang, D.; Lin, Y.; Xie, T. Photoelectrical properties of CdS/CdSe core/shell QDs modified anatase TiO2 nanowires and their application for solar cells. Phys. Chem. Chem. Phys. 2017, 19, 15724–15733. [Google Scholar] [CrossRef]
- Moon, H.; Lee, C.; Lee, W.; Kim, J.; Chae, H. Stability of Quantum Dots, Quantum Dot Films, and Quantum Dot Light-Emitting Diodes for Display Applications. Adv. Mater. 2019, 31, e1804294. [Google Scholar] [CrossRef]
- Matos, B.; Martins, M.; Samamed, A.C.; Sousa, D.; Ferreira, I.; Diniz, M.S. Toxicity Evaluation of Quantum Dots (ZnS and CdS) Singly and Combined in Zebrafish (Danio rerio). Int. J. Environ. Res. Public Health 2019, 17, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modlitbová, P.; Pořízka, P.; Novotný, K.; Drbohlavová, J.; Chamradová, I.; Farka, Z.; Zlámalová-Gargošová, H.; Romih, T.; Kaiser, J. Short-term assessment of cadmium toxicity and uptake from different types of Cd-based Quantum Dots in the model plant Allium cepa L. Ecotoxicol. Environ. Saf. 2018, 153, 23–31. [Google Scholar] [CrossRef]
- Favero, P.P.; de Souza-Parise, M.; Fernandez, J.L.R.; Miotto, R.; Ferraz, A.C. Surface properties of CdS nanoparticles. Braz. J. Phys. 2006, 36, 1032–1034. [Google Scholar] [CrossRef]
- Wu, T.; Tang, M. Toxicity of quantum dots on respiratory system. Inhal. Toxicol. 2014, 26, 128–139. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pagano, L.; Sardaro, M.L.S.; Villani, M. Genome-Wide Approach in Arabidopsis thaliana to Assess the Toxicity of Cadmium Sulfide Quantum Dots. Environ. Sci. Technol. 2014, 48, 5902–5909. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, R.; Maestri, E.; Pagano, L.; Marmiroli, M.; White, J.C.; Marmiroli, N. Plant Response to Metal-Containing Engineered Nanomaterials: An Omics-Based Perspective. Environ. Sci. Technol. 2018, 52, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Maestri, E.; Caldara, M.; White, J.C.; Marmiroli, N.; Marmiroli, M. Engineered Nanomaterial Activity at the Organelle Level: Impacts on the Chloroplasts and Mitochondria. ACS Sustain. Chem. Eng. 2018, 6, 12562–12579. [Google Scholar] [CrossRef]
- Pasquali, F.; Agrimonti, C.; Pagano, L.; Zappettini, A.; Villani, M.; Marmiroli, M.; White, J.C.; Marmiroli, N. Nucleo-mitochondrial interaction of yeast in response to cadmium sulfide quantum dot exposure. J. Hazard. Mater. 2017, 324, 744–752. [Google Scholar] [CrossRef]
- Paesano, L.; Marmiroli, M.; Bianchi, M.G.; White, J.C.; Bussolati, O.; Zappettini, A.; Villani, M.; Marmiroli, N. Differences in toxicity, mitochondrial function and miRNome in human cells exposed in vitro to Cd as CdS quantum dots or ionic Cd. J. Hazard. Mater. 2020, 393, 122430. [Google Scholar] [CrossRef]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta-analysis of cellular toxicity for cadmi-um-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Gong, J.; Chen, J.; Zeng, Z.; Huang, W.; Pu, K.; Liu, J.; Chen, P. Recent advances on Graphene Quantum Dots: From chemistry and physics to applications. Adv. Mater. 2019, 31, e1808283. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, M. Dysfunction of various organelles provokes multiple cell death after quantum dot exposure. Int. J. Nanomed. 2018, 13, 2729–2742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, T.L.; Bilbao, E.; Cardoso, C.; Soto, M.; Bebianno, M.J. Changes in metallothionein transcription levels in the mussel Mytilus galloprovincialis exposed to CdTe quantum dots. Ecotoxicology 2018, 27, 402–410. [Google Scholar] [CrossRef]
- Majumdar, S.; Ma, C.; Villani, M.; Zuverza-Mena, N.; Pagano, L.; Huang, Y.; Zappettini, A.; Keller, A.A.; Marmiroli, N.; Dhankher, O.P.; et al. Surface coating determines the response of soybean plants to cadmium sulfide quantum dots. NanoImpact 2019, 14, 100151. [Google Scholar] [CrossRef]
- Marmiroli, M.; Mussi, F.; Pagano, L.; Imperiale, D.; Lencioni, G.; Villani, M.; Zappettini, A.; White, J.C.; Marmiroli, N. Cadmium sulfide quantum dots impact Arabidopsis thaliana physiology and morphology. Chemosphere 2020, 240, 124856. [Google Scholar] [CrossRef]
- Marmiroli, M.; Imperiale, D.; Pagano, L.; Villani, M.; Zappettini, A.; Marmiroli, N. The proteomic response of Arabidopsis thaliana to cadmium sulfide quantum dots, and its correlation with the transcriptomic response. Front. Plant Sci. 2015, 6, 1104. [Google Scholar] [CrossRef] [Green Version]
- Marmiroli, M.; Lepore, G.O.; Pagano, L.; D’Acapito, F.; Gianoncelli, A.; Villani, M.; Lazzarini, L.; White, J.C.; Marmiroli, N. The fate of CdS quantum dots in plants as revealed by extended X-ray absorption fine structure (EXAFS) analysis. Environ. Sci. Nano 2020, 7, 1150–1162. [Google Scholar] [CrossRef]
- Hajduch, M.; Hearne, L.B.; Miernyk, J.A.; Casteel, J.E.; Joshi, T.; Agrawal, G.K.; Song, Z.; Zhou, M.; Xu, D.; Thelen, J.J. Systems Analysis of Seed Filling in Arabidopsis: Using General Linear Modeling to Assess Concordance of Transcript and Protein Expression. Plant Physiol. 2010, 152, 2078–2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pütz, S.M.; Boehm, A.M.; Stiewe, T.; Sickmann, A. iTRAQ Analysis of a Cell Culture Model for Malignant Transformation, Including Comparison with 2D-PAGE and SILAC. J. Proteome Res. 2012, 11, 2140–2153. [Google Scholar] [CrossRef]
- Gallo, V.; Srivastava, V.; Bulone, V.; Zappettini, A.; Villani, M.; Marmiroli, N.; Marmiroli, M. Proteomic analysis identifies markers of exposure to Cadmium Sulfide Quantum Dots (CdS QDs). Nanomaterials 2020, 10, 1214. [Google Scholar] [CrossRef]
- Rocco, M.; D’Ambrosio, C.; Arena, S.; Faurobert, M.; Scaloni, A.; Marra, M. Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics 2006, 6, 3781–3791. [Google Scholar] [CrossRef]
- Faurobert, M.; Mihr, C.; Bertin, N.; Pawlowski, T.; Negroni, L.; Sommerer, N.; Causse, M. Major Proteome Variations Associated with Cherry Tomato Pericarp Development and Ripening. Plant Physiol. 2007, 143, 1327–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramagli, L.S.; Rodriguez, L.V. Quantitation of microgram amounts of protein in SDS -mercaptoethanol-tris electrophoresis sample buffer. Electrophoresis 1985, 6, 559–563. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M.J. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identifi-cation and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 en-codes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Jha, S.K.; Bagri, J. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tol-erance in Arabidopsis. PLoS ONE 2015, 10, e0125168. [Google Scholar]
- Andriotis, V.M.E.; Kruger, N.J.; Pike, M.J.; Smith, A.M. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 2009, 185, 649–662. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, S.-L.; Xie, L.-F.; Puah, C.S.; Zhang, X.-Q.; Yang, W.-C.; Sundaresan, V.; Ye, D. VANGUARD1 Encodes a Pectin Methylesterase That Enhances Pollen Tube Growth in the Arabidopsis Style and Transmitting Tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef] [Green Version]
- Semane, B.; Dupae, J.; Cuypers, A.; Noben, J.P.; Tuomainen, M.; Karenlampi, S.; Van Belleghem, F.; Smeets, K.; Vangrosveld, J. Leaf proteome responses of Arabidopsis thaliana exposed to mild Cadmium stress. J.Plant Physiol. 2010, 167, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Villalobos, L.I.A.; Nill, C.; Marrocco, K.; Kretsch, T.; Schwechheimer, C. The evolutionarily conserved Ara-bidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 2007, 392, 106–116. [Google Scholar] [CrossRef]
- Yi, X.; McChargue, M.; Laborde, S.; Frankel, L.K.; Bricker, T.M. The manganese-stabilizing protein is required for pho-tosystem II assembly/stability and photoautotrophy in higher plants. J. Biol. Chem. 2005, 280, 16170–16174. [Google Scholar] [CrossRef] [Green Version]
- Kosma, D.K.; Molina, I.; Ohlrogge, J.B.; Pollard, M. Identification of an Arabidopsis Fatty Alcohol:Caffeoyl-Coenzyme A Acyltransferase Required for the Synthesis of Alkyl Hydroxycinnamates in Root Waxes. Plant Physiol. 2012, 160, 237–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perales, M.; Eubel, H.; Heinemeyer, J.; Colaneri, A.; Zabaleta, E.; Braun, H.-P. Disruption of a Nuclear Gene Encoding a Mitochondrial Gamma Carbonic Anhydrase Reduces Complex I and Supercomplex I+III2 Levels and Alters Mitochondrial Physiology in Arabidopsis. J. Mol. Biol. 2005, 350, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Briefings Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bach-Pages, M.; Homma, F.; Kourelis, J.; Kaschani, F.; Mohammed, S.; Kaiser, M.; Van Der Hoorn, R.A.L.; Castello, A.; Preston, G.M. Discovering the RNA-Binding Proteome of plant leaves with an improved RNA interactome capture method. Biomolecules 2020, 10, 661. [Google Scholar] [CrossRef] [PubMed]
- Sarry, J.-E.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D.; et al. The early responses ofArabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef]
- Kwak, K.J.; Kim, Y.O.; Kang, H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005, 56, 3007–3016. [Google Scholar] [CrossRef]
- Lawere, S.L.; Raskar, S. Effect of titanium dioxide nanoparticles on hydrolytic and antioxidant enzymes during seed germination in onion. Int. J. Curr. Micorobiol. App. Sci. 2014, 3, 749–760. [Google Scholar]
- Ueda, T.; Yamaguchi, M.; Uchimiya, H.; Nakano, A. Ara6, a plant-unique novel type Rab GTPase, functions in the endo-cytic pathway of Arabidopsis thaliana. EMBO J. 2001, 20, 4730–4741. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, M.; Fermani, S.; Costa, A.; Lemaire, S.D.; Trost, P. Plant cytoplasmic GAPDH: Redox post-translational modi-fications and moonlighting properties. Front. Plant Sci. 2013, 4, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez-Casati, D.F. Characterization of Arabidopsis Lines Deficient in GAPC-1, a Cytosolic NAD-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase. Plant Physiol. 2008, 148, 1655–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baalmann, E.; Backhausen, J.E.; Rak, C.; Vetter, S.; Scheibe, R. Reductive modification and nonreductive activation of purified spinach chloroplast NADP-dependent glyceraldehyde-3-phosphate dehydrogenase. Arch. Biochem. Biophys. 1995, 324, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Price, G.D.; Evans, J.; Caemmerer, S.; Yu, J.-W.; Badger, M. Specific reduction of chloroplast glyceraldehyde-3-phosphate dehydrogenase activity by antisense RNA reduces CO2 assimilation via a reduction in ribulose bisphosphate regeneration in transgenic tobacco plants. Planta 1995, 195, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Garcìa-Sànchez, S.; Bernales, I.; Cristobal, S. Early response to nanoparticles in the Arabidopsis transcriptome compromises plant defence and root-hair development through salicylic acid signaling. BMC Genom. 2015, 16, 341. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, M.; Krishnamurthy, S.; Shukla, D.; Kiiskila, J.; Jain, A.; Datta, R.; Sharma, N.; Sahi, S.V. Comparative transcriptome and proteome analysis to reveal the biosynthesis of gold nanoparticles in Arabidopsis. Sci. Rep. 2016, 6, 21733. [Google Scholar] [CrossRef] [Green Version]
- Marmiroli, M.; Maestri, E.; Pagano, L.; Robinson, B.H.; Ruotolo, R.; Marmiroli, N. Toxicology assessment of engineered nanomaterials: Innovation and tradition. In Exposure to Engineered Nanomaterials in the Environment; Elsevier BV: Amsterdam, The Netherlands, 2019; Volume 8, pp. 209–234. [Google Scholar]
- Giegé, P.; Heazlewood, J.L.; Roessner-Tunali, U.; Millar, A.H.; Fernie, A.R.; Leaver, C.J.; Sweetlove, L.J. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 2003, 15, 2140–2151. [Google Scholar] [CrossRef] [Green Version]
- Barkla, B.J.; Vera-Estrella, R.; Hernández-Coronado, M.; Pantoja, O. Quantitative proteomics of the tonoplast Reveals a Role for Glycolytic Enzymes in Salt Tolerance. Plant Cell 2009, 21, 4044–4058. [Google Scholar] [CrossRef] [Green Version]
- Hossain, Z.; Mustafa, G.; Komatsu, S. Plant Responses to Nanoparticle Stress. Int. J. Mol. Sci. 2015, 16, 26644–26653. [Google Scholar] [CrossRef] [Green Version]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the Diversity of the Arabidopsis glutathione S-Transferase Gene Family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Glutathione Transferases. Arab. Book 2010, 8, e0131. [Google Scholar] [CrossRef] [Green Version]
- Moons, A. Regulatory and Functional Interactions of Plant Growth Regulators and Plant Glutathione S-Transferases (GSTs). Vitam. Horm. 2005, 72, 155–202. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.M.G.; Park, S.Y.; Choi, J. Evaluation of the effect of silver nanoparticles and silver ions using stress responsive gene expression in Chironomus riparius. Chemosphere 2013, 92, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Sharma, N.C.; Fleischmann, P.; Burbage, J.; Venkatachalam, P.; Sahi, S.V. Nanotitania Exposure Causes Alterations in Physiological, Nutritional and Stress Responses in Tomato (Solanum lycopersicum). Front. Plant Sci. 2017, 8, 633. [Google Scholar] [CrossRef] [Green Version]
- Pagano, L.; Pasquali, F.; Majumdar, S.; De La Torre-Roche, R.; Zuverza-Mena, N.; Villani, M.; Zappettini, A.; Marra, R.E.; Isch, S.M.; Marmiroli, M.; et al. Exposure of Cucurbita pepo to binary combinations of engineered nanomaterials: Physiological and molecular response. Environ. Sci. Nano 2017, 4, 1579–1590. [Google Scholar] [CrossRef]
- Tang, Y.; He, R.; Zhao, J.; Nie, G.; Xu, L.; Xing, B. Oxidative stress-induced toxicity of CuO nanoparticles and related toxicogenomic responses in Arabidopsis thaliana. Environ. Pollut. 2016, 212, 605–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landa, P.; Prerostova, S.; Petrova, S.; Knirsch, V.; Vankova, R.; Vanek, T. The transcriptomic response of Arabidopsis thaliana to Zinc Oxide: A comparison of the impact of Nanoparticle, Bulk, and Ionic Zinc. Environ. Sci. Technol. 2015, 49, 14537–14545. [Google Scholar] [CrossRef]
- Syu, Y.-Y.; Hung, J.-H.; Chen, J.-C.; Chuang, H.-W. Impacts of size and shape of silver nanoparticles on Arabidopsis plant growth and gene expression. Plant Physiol. Biochem. 2014, 83, 57–64. [Google Scholar] [CrossRef]
- De Smet, I.; Signora, L.; Beeckman, T.; Inzé, D.; Foyer, C.H.; Zhang, H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003, 33, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Chandler, P.M.; Robertson, M. Gene Expression Regulated by Abscisic Acid and its Relation to Stress Tolerance. Annu. Rev. Plant Biol. 1994, 45, 113–141. [Google Scholar] [CrossRef]
- Jaillais, Y.; Chory, J. Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 2010, 17, 642–645. [Google Scholar] [CrossRef] [Green Version]
- Mehlgarten, C.; Prochaska, H.; Hammermeister, A.; Abdel-Fattah, W.; Wagner, M.; Krutyhołowa, R.; Jun, S.E.; Kim, G.-T.; Glatt, S.; Breunig, K.D.; et al. Use of a Yeast tRNase Killer Toxin to Diagnose Kti12 Motifs Required for tRNA Modification by Elongator. Toxins 2017, 9, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woloszynska, M.; Le Gall, S.; Van Lijsebettens, M. Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 1025–1033. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, H.; Jablonowski, D.; Zhou, X.; Ren, X.; Hong, X.; Schaffrath, R.; Zhu, J.-K.; Gong, Z. Mutations in ABO1/ELO2, a Subunit of Holo-Elongator, Increase Abscisic Acid Sensitivity and Drought Tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006, 26, 6902–6912. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Hua, D.; Chen, Z.; Zhou, Z.; Gong, Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009, 60, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Huang, W.; Li, Y.; Wang, H.; Huang, H.; Cui, X. Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J. 2011, 69, 792–808. [Google Scholar] [CrossRef]
- DeFraia, C.T.; Wang, Y.; Yao, J.; Mou, Z. Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 2013, 13, 102. [Google Scholar] [CrossRef] [Green Version]
- Rissler, H.M.; Collakova, E.; Della Penna, D.; Whelan, J.; Pogson, B.J. Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 2002, 128, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-S.; Li, H.-M. Arabidopsis CHLI2 Can Substitute for CHLI1. Plant Physiol. 2009, 150, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Arimura, S.-I.; Kurisu, R.; Sugaya, H.; Kadoya, N.; Tsutsumi, N. Cold Treatment Induces Transient Mitochondrial Fragmentation in Arabidopsis thaliana in a Way that Requires DRP3A but not ELM1 or an ELM1-Like Homologue, ELM2. Int. J. Mol. Sci. 2017, 18, 2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couturier, J.; Touraine, B.; Briat, J.F.; Gaymard, F.; Rouhier, N. The iron-sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front. Plant Sci. 2013, 4, 259. [Google Scholar] [CrossRef] [Green Version]
- Bayer, E.M.; Bottrill, A.R.; Walshaw, J.; Vigouroux, M.; Naldrett, M.J.; Thomas, C.L.; Maule, A.J. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 2006, 6, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-F.; Lin, K.-H.; He, S.-L.; Chen, J.-L.; Jiang, J.-Z.; Chen, B.-H.; Hou, Y.-S.; Chen, R.-S.; Hong, C.-Y.; Ho, S.-L. Multiple Patterns of Regulation and Overexpression of a Ribonuclease-Like Pathogenesis-Related Protein Gene, OsPR10a, Conferring Disease Resistance in Rice and Arabidopsis. PLoS ONE 2016, 11, e0156414. [Google Scholar] [CrossRef] [PubMed]
- Mo, P.; Zhu, Y.; Liu, X.; Zhang, A.; Yan, C.; Wang, D. Identification of two phosphatidylinositol/phosphatidylcholine transfer protein genes that are predominately transcribed in the flowers of Arabidopsis thaliana. J. Plant Physiol. 2007, 164, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Capelli, N.; Diogon, T.; Greppin, H.; Simon, P. Isolation and characterization of a cDNA clone encoding an osmotin-like protein from Arabidopsis thaliana. Gene 1997, 191, 51–56. [Google Scholar] [CrossRef]
- Tumburu, L.; Andersen, C.P.; Rygiewicz, P.T.; Reichman, J.R. Molecular and Physiological Responses to Titanium dioxide and Cerium Oxide nanoparticles in Arabidopsis. Environ. Toxicol. Chem. 2017, 36, 71–82. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallo, V.; Zappettini, A.; Villani, M.; Marmiroli, N.; Marmiroli, M. Comparative Analysis of Proteins Regulated during Cadmium Sulfide Quantum Dots Response in Arabidopsis thaliana Wild Type and Tolerant Mutants. Nanomaterials 2021, 11, 615. https://doi.org/10.3390/nano11030615
Gallo V, Zappettini A, Villani M, Marmiroli N, Marmiroli M. Comparative Analysis of Proteins Regulated during Cadmium Sulfide Quantum Dots Response in Arabidopsis thaliana Wild Type and Tolerant Mutants. Nanomaterials. 2021; 11(3):615. https://doi.org/10.3390/nano11030615
Chicago/Turabian StyleGallo, Valentina, Andrea Zappettini, Marco Villani, Nelson Marmiroli, and Marta Marmiroli. 2021. "Comparative Analysis of Proteins Regulated during Cadmium Sulfide Quantum Dots Response in Arabidopsis thaliana Wild Type and Tolerant Mutants" Nanomaterials 11, no. 3: 615. https://doi.org/10.3390/nano11030615
APA StyleGallo, V., Zappettini, A., Villani, M., Marmiroli, N., & Marmiroli, M. (2021). Comparative Analysis of Proteins Regulated during Cadmium Sulfide Quantum Dots Response in Arabidopsis thaliana Wild Type and Tolerant Mutants. Nanomaterials, 11(3), 615. https://doi.org/10.3390/nano11030615